| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,945,708 – 26,945,800 |
| Length | 92 |
| Max. P | 0.703018 |

| Location | 26,945,708 – 26,945,800 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 59.19 |
| Shannon entropy | 0.80287 |
| G+C content | 0.54814 |
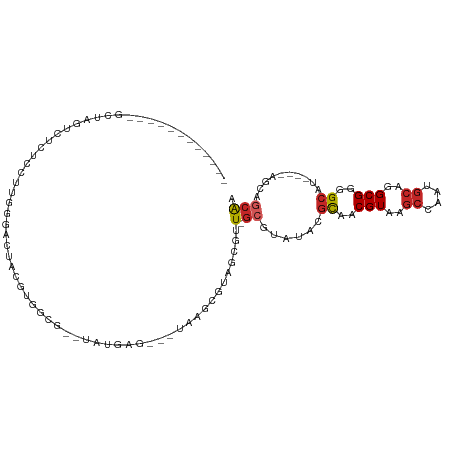
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -7.99 |
| Energy contribution | -8.05 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
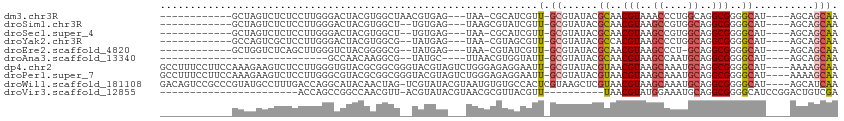
>dm3.chr3R 26945708 92 - 27905053 ------------GCUAGUCUCUCCUUGGGACUACGUGGCUAACGUGAG---UAA-CGCAUCGUU-GCGUAUACGCAACGUAAACCCUGGCAGGCGGGGCAU----AGCAGCAA ------------(((((((((.(((.(((..(((((.((..((....)---).(-((((....)-))))....)).)))))..)))....))).))))).)----)))..... ( -36.80, z-score = -2.34, R) >droSim1.chr3R 26586406 91 - 27517382 ------------GCUAGUCUCUCCUUGGGACUACGUGGCU--UGUGAG---UAAGCGUAUCGUU-GCGUAUACGCAACGUAAGCCGUGGCAGGCGGGGCAU----AGCAGCAA ------------(((((((((.(((...(.(((((..(((--((....---)))))((..((((-(((....)))))))...))))))))))).))))).)----)))..... ( -34.60, z-score = -1.06, R) >droSec1.super_4 5763986 90 - 6179234 ------------GCUAGUCUCUCCUUGGGACUACGUGGCU--UGUGAG---UAA-CGCAUCGUU-GCGUAUACGCAACGUAAGCCGUGGCAGGCGGGGCAU----AGCAGCAA ------------(((((((((.(((...(.((((..((((--((((..---...-)))..((((-(((....))))))).))))))))))))).))))).)----)))..... ( -35.20, z-score = -1.40, R) >droYak2.chr3R 27879102 90 - 28832112 ------------GCCAGUCGCUCCUUGGGACUACGUGGCG--UAUGAG---UAA-CGUAGCGUU-GCGUAUACGCCACGUAAGCCCUGGCAGGCGGGGCAU----AGCAGCAA ------------(((...((((((..(((..(((((((((--((((.(---(((-((...))))-)).)))))))))))))..))).))..)))).)))..----........ ( -46.60, z-score = -3.95, R) >droEre2.scaffold_4820 9428219 89 + 10470090 ------------GCUGGUCUCAGCUUGGGUCUACGGGGCG--UAUGAG---UAA-CGUAUCGUU-GCGUAUACGCAACGUAAGCCCU-GCAGGCGGGGCAU----AGCAGCAA ------------(((((((((.(((((((..((((..(((--((((.(---(((-((...))))-)).)))))))..))))..))).-..))))))))).)----)))..... ( -44.00, z-score = -4.48, R) >droAna3.scaffold_13340 14326068 74 - 23697760 ----------------------------GCCAACAAGGCG--UAUGC----UUAACGUGGUAUU-GCGUAUACGCAACGUAAGCCAAUGCAGGCGGGGCAU----AGCAGCAA ----------------------------(((.....))).--(((((----(...(((.(((((-(.((.((((...)))).))))))))..))).)))))----)....... ( -23.90, z-score = -0.38, R) >dp4.chr2 30598506 108 + 30794189 GCCUUUCCUUCCAAAGAAGUCUCCUUGGGUGUACGCGGCGGGUACGUAGUCUGGGAGAGGAAUU-GCGUAUACGUAACGUAAGCAAAUGCAGGCGGGGCAU----AAAAGCAA ((((..(((.((((.((....)).))))((.((((..(((..((((((((((......)).)))-)))))..)))..)))).))......)))..))))..----........ ( -35.50, z-score = -1.18, R) >droPer1.super_7 1464029 108 + 4445127 GCCUUUCCUUCCAAAGAAGUCUCCUUGGGCGUACGCGGCGGGUACGUAGUCUGGGAGAGGAAUU-GCGUAUACGUAACGUAAGCAAAUGCAGGCGGGGCAU----AAAAGCAA ((((..(((.((((.((....)).))))((.((((..(((..((((((((((......)).)))-)))))..)))..)))).))......)))..))))..----........ ( -37.00, z-score = -1.39, R) >droWil1.scaffold_181108 604049 108 - 4707319 GACAGUCCGCCCGUAUGCCUUUGACCAGGCAUACAACUAG-UCGUAUACGUAAUGUGUGCCACUCGUAAGCUCGUAACGUAAGCAAAUGCAGGCGGGGCAU----AGCAUCAA ........((((((((((((......)))))))).((.((-(.((((((.....)))))).))).))..........(((..((....))..)))))))..----........ ( -34.60, z-score = -1.45, R) >droVir3.scaffold_12855 8786165 79 + 10161210 -----------------------ACCAGCCGGCCAACGUU-ACGUAUACGUAACGCGUUACGUU----------UAACGUAUGGAAAUGCAGGCGGGGCAUCCGGACUGUCGA -----------------------..(((((((....((((-(((....))))))).((..((((----------(...((((....)))))))))..))..)))).))).... ( -27.80, z-score = -1.05, R) >consensus ____________GCUAGUCUCUCCUUGGGACUACGUGGCG__UAUGAG___UAAGCGUAGCGUU_GCGUAUACGCAACGUAAGCCAAUGCAGGCGGGGCAU____AGCAGCAA .........................................................................((..(((..((....))..)))..)).............. ( -7.99 = -8.05 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:53 2011