
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,286,419 – 9,286,520 |
| Length | 101 |
| Max. P | 0.896950 |

| Location | 9,286,419 – 9,286,520 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.60 |
| Shannon entropy | 0.53239 |
| G+C content | 0.48747 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -11.87 |
| Energy contribution | -12.02 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9286419 101 + 23011544 ------CGCACACACACACACACACCUUGUUGCAGAUGAGAAAGGGCACAUGUGUGUGUGUGUGAGGGUGAAAUGAGCUAGAAAUUAGUUUAUGGCGAAGGUCGUGC ------.((((.(((((((((((((..(((.((...(.....)..))))).)))))))))))))........(((((((((...)))))))))(((....))))))) ( -40.30, z-score = -4.09, R) >droSim1.chr2L 9063154 99 + 22036055 ------CACACACACACACACACACCUUGAUCCGGAUGAGAACGGGCACAUGUGUGUGUGUG--AGGGUGAAAUGAGCUAGAAAUUAGUUUAUGGCGAAGGUCGUGC ------(((.(.((((((((((((...((.((((........))))))..))))))))))))--.).)))..(((((((((...)))))))))(((....))).... ( -38.30, z-score = -3.82, R) >droSec1.super_3 4745286 99 + 7220098 ------CACUCACACACACACACACCUUGAUCCGGAUGAGAACGUGCACAUGUGUGUGUGUG--AGGGUGAAGUGAGCUAGAAAUUAGUUUAUGGCGAAGGUCGUGC ------(((((.((((((((((((...((...((........))..))..))))))))))))--.)))))..(((((((((...)))))))))(((....))).... ( -37.90, z-score = -2.97, R) >droEre2.scaffold_4929 9893539 103 + 26641161 CACUCGCACACACACUAACACACACCCUUAUCCGGAUGAGAACGUGCAUAUGUGUGUGUGGG----GGUGAAAUGAGCUAGAAAUUAGUUUACGGCGAAGGUCGUGC (((((.(((((((((.....(((...(((((....)))))...))).....))))))))).)----))))....(((((((...)))))))(((((....))))).. ( -39.60, z-score = -3.78, R) >droAna3.scaffold_12943 626561 95 + 5039921 --AACACACAUACACUCACGUACUUAUCCUCACGAGCGUGUGAGUAGGGGUUUGUCAGUGAA----------AUGAGCUAGAAAUUAGUUUAUGACGAAGGUCGUCC --..........((((.(((.((((...((((((....))))))...)))).))).))))..----------(((((((((...)))))))))(((....))).... ( -25.90, z-score = -1.17, R) >dp4.chr4_group5 1610348 86 + 2436548 ------CACCAACACCUACACAC--ACCUCCACCGAUGAGAGAGAGAGCG---GGCUGUGUA----------AUGAGCUAGAAAUUAGUUUAUGACGAAGGUCGUCC ------..........((((((.--.((.((.((.......).).).).)---)..))))))----------(((((((((...)))))))))(((....))).... ( -17.30, z-score = 0.04, R) >droPer1.super_5 6018875 86 + 6813705 ------CACCAACACCUACACAC--ACCUCCACCGAUGAGAGAGAGAGCG---GGCUGUGUA----------AUGAGCUAGAAAUUAGUUUAUGACGAAGGUCGUCC ------..........((((((.--.((.((.((.......).).).).)---)..))))))----------(((((((((...)))))))))(((....))).... ( -17.30, z-score = 0.04, R) >consensus ______CACACACACACACACACACCUUGAUACGGAUGAGAAAGUGCACAUGUGUGUGUGUG____GGUGAAAUGAGCUAGAAAUUAGUUUAUGGCGAAGGUCGUGC ...............(((((.((((..(((...............)))...)))).)))))...........(((((((((...)))))))))(((....))).... (-11.87 = -12.02 + 0.15)

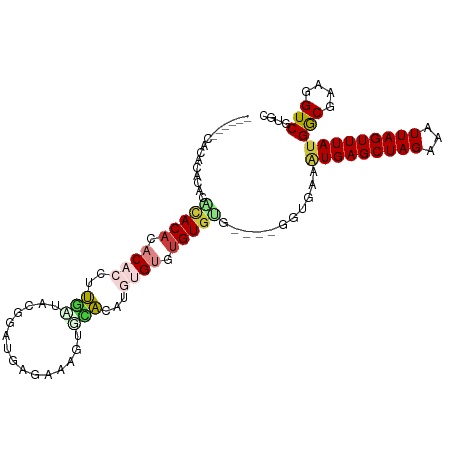
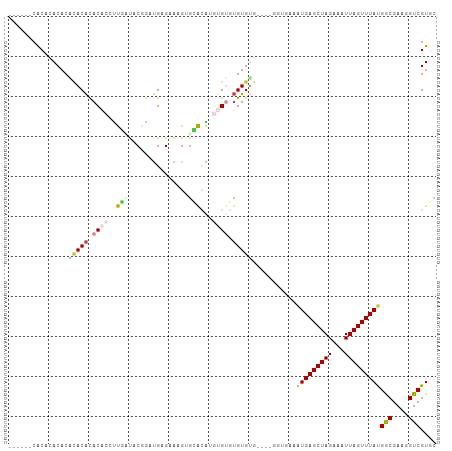
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:56 2011