
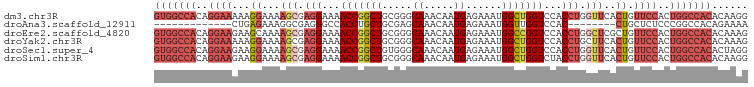
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,812,875 – 26,812,974 |
| Length | 99 |
| Max. P | 0.892657 |

| Location | 26,812,875 – 26,812,974 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.70 |
| Shannon entropy | 0.31884 |
| G+C content | 0.55193 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -21.32 |
| Energy contribution | -23.77 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
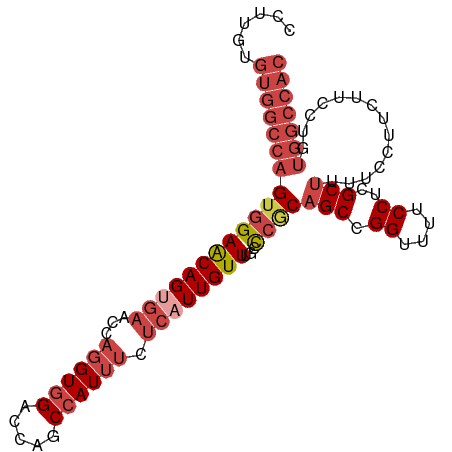
>dm3.chr3R 26812875 99 + 27905053 CCUUGUGUGGCCAGUGGAACAGUGAACCAGGUGGACCAGCCAUUUCUCAUUGUUUGCCCGCAGCCGGUUUUCCUCGCUUUUCCUUUUUCCUGUGGCCAC ......(((((((((((((((((((...((((((.....)))))).))))))))...))))(((.((....))..)))..............))))))) ( -31.60, z-score = -1.47, R) >droAna3.scaffold_12911 1237325 78 - 5364042 UUUUCUGUGGCCGGGAGAGCAG--------GUGGACCAACCAUUUCUCAUUGUUUGCUCGCAGCAGGUGGCCCUCGCCUUUCUCAG------------- ....(((.(((.(((.(((.((--------((((.....))))))))).(..(((((.....)))))..))))..))).....)))------------- ( -23.70, z-score = 0.06, R) >droEre2.scaffold_4820 9296796 99 - 10470090 CUUUGUGUGGCCAGUGGAACAGCGAGCCAGGUGGACCGGCCAUUUCUCAUUGUUUGCCCGCAGCCGGUUUUCCUCGCUUUUGCUUCUUCCUGUGGCCAC ......(((((((..((((.(((((((.(((..(((((((.......((.....))......)))))))..))).))))..)))..))))..))))))) ( -41.52, z-score = -3.27, R) >droYak2.chr3R 27748295 99 + 28832112 CUUUGUGUGGCCAGUGGAACAGUGAAGCAGGUGGACCAGCCAUUUCUCAUUGUUUGCCCGCAGCCGGUUUUCCUCGCUUUUCCUUUUUCCUGUGGCCAC ......(((((((..((((.((.((((((((..((((.((.......((.....))......)).))))..))).)))))..))..))))..))))))) ( -32.82, z-score = -1.63, R) >droSec1.super_4 5637852 99 + 6179234 CCUAGUGUGGCCAGUGGAACAGUGAACCAGGUGGACCAGCCAUUUCUCAUUGUUUGCCCACAGCCGGUUUUCCUCGCUUUUCCUUCUUCCUGUGGCCAC ......(((((((((((((((((((...((((((.....)))))).))))))))...))))(((.((....))..)))..............))))))) ( -32.00, z-score = -1.62, R) >droSim1.chr3R 26457362 99 + 27517382 CCUUGUGUGGCCAGUGGAACAGUGAACCAGGUAGACCAGCCAUUUCUCAUUGUUUGCCCGCAGCCGGUUUUCCUCGCUUUUCCUUCUUCCUGUGGCCAC ......(((((((((((((((((((....(((......))).....))))))))...))))(((.((....))..)))..............))))))) ( -29.40, z-score = -1.28, R) >consensus CCUUGUGUGGCCAGUGGAACAGUGAACCAGGUGGACCAGCCAUUUCUCAUUGUUUGCCCGCAGCCGGUUUUCCUCGCUUUUCCUUCUUCCUGUGGCCAC ......(((((((((((((((((((...((((((.....)))))).))))))))...))))(((.((....))..)))..............))))))) (-21.32 = -23.77 + 2.45)


| Location | 26,812,875 – 26,812,974 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.70 |
| Shannon entropy | 0.31884 |
| G+C content | 0.55193 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -23.95 |
| Energy contribution | -26.62 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
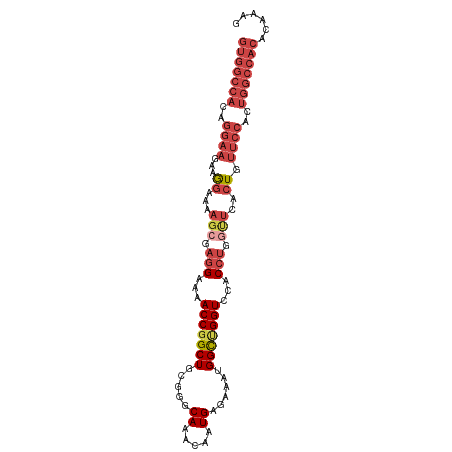
>dm3.chr3R 26812875 99 - 27905053 GUGGCCACAGGAAAAAGGAAAAGCGAGGAAAACCGGCUGCGGGCAAACAAUGAGAAAUGGCUGGUCCACCUGGUUCACUGUUCCACUGGCCACACAAGG (((((((..((((...((...(((.(((...(((((((.....((.....))......)))))))...))).)))..)).))))..)))))))...... ( -35.00, z-score = -1.83, R) >droAna3.scaffold_12911 1237325 78 + 5364042 -------------CUGAGAAAGGCGAGGGCCACCUGCUGCGAGCAAACAAUGAGAAAUGGUUGGUCCAC--------CUGCUCUCCCGGCCACAGAAAA -------------(((.(...((.((((((....(((.....)))..((((.(....).))))......--------..))))))))...).))).... ( -20.60, z-score = 0.46, R) >droEre2.scaffold_4820 9296796 99 + 10470090 GUGGCCACAGGAAGAAGCAAAAGCGAGGAAAACCGGCUGCGGGCAAACAAUGAGAAAUGGCCGGUCCACCUGGCUCGCUGUUCCACUGGCCACACAAAG (((((((..((((..(((...(((.(((...(((((((.....((.....))......)))))))...))).))).))).))))..)))))))...... ( -42.20, z-score = -3.56, R) >droYak2.chr3R 27748295 99 - 28832112 GUGGCCACAGGAAAAAGGAAAAGCGAGGAAAACCGGCUGCGGGCAAACAAUGAGAAAUGGCUGGUCCACCUGCUUCACUGUUCCACUGGCCACACAAAG (((((((..((((...((..((((.(((...(((((((.....((.....))......)))))))...)))))))..)).))))..)))))))...... ( -36.90, z-score = -2.84, R) >droSec1.super_4 5637852 99 - 6179234 GUGGCCACAGGAAGAAGGAAAAGCGAGGAAAACCGGCUGUGGGCAAACAAUGAGAAAUGGCUGGUCCACCUGGUUCACUGUUCCACUGGCCACACUAGG (((((((..((((..((....(((.(((...(((((((((...((.....))....)))))))))...))).)))..)).))))..)))))))...... ( -37.60, z-score = -2.38, R) >droSim1.chr3R 26457362 99 - 27517382 GUGGCCACAGGAAGAAGGAAAAGCGAGGAAAACCGGCUGCGGGCAAACAAUGAGAAAUGGCUGGUCUACCUGGUUCACUGUUCCACUGGCCACACAAGG (((((((..((((..((....(((.(((...(((((((.....((.....))......)))))))...))).)))..)).))))..)))))))...... ( -35.00, z-score = -1.99, R) >consensus GUGGCCACAGGAAGAAGGAAAAGCGAGGAAAACCGGCUGCGGGCAAACAAUGAGAAAUGGCUGGUCCACCUGGUUCACUGUUCCACUGGCCACACAAAG (((((((..((((...((...(((.(((...(((((((.....((.....))......)))))))...))).)))..)).))))..)))))))...... (-23.95 = -26.62 + 2.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:44 2011