| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,787,049 – 26,787,202 |
| Length | 153 |
| Max. P | 0.833677 |

| Location | 26,787,049 – 26,787,202 |
|---|---|
| Length | 153 |
| Sequences | 10 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 51.97 |
| Shannon entropy | 1.00040 |
| G+C content | 0.51591 |
| Mean single sequence MFE | -58.38 |
| Consensus MFE | -0.63 |
| Energy contribution | -0.83 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
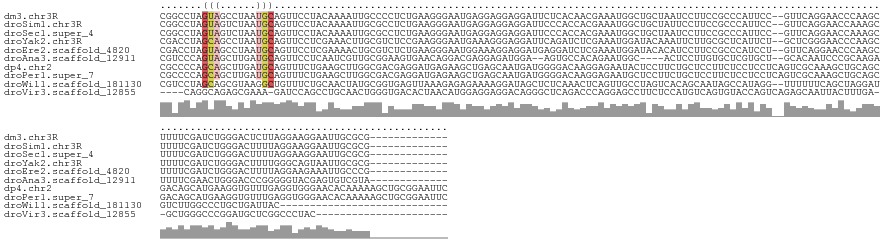
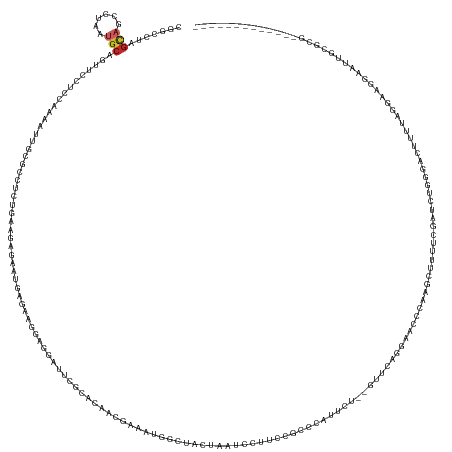
>dm3.chr3R 26787049 153 + 27905053 CGGCCUAGUAGCCUAAUGCAGUUCCUACAAAAUUGCCCCUCUGAAGGGAAUGAGGAGGAGGAUUCUCACAACGAAAUGGCUGCUAAUCCUUCCGCCCAUUCC--GUUCAGGAACCCAAGCUUUUCGAUCUGGGACUCUUAGGAAGGAAUUGCGCG------------- .(((......)))....(((((((((.(...........((((((.((((((.((.((((((((..((((......))..))..))))).))).))))))))--.))))))..((((..(.....)...))))........).)))))))))...------------- ( -50.90, z-score = -1.36, R) >droSim1.chr3R 26431359 153 + 27517382 CGGCCUAGUAGUCUAAUGCAGUUCCUACAAAAUUGCGCCUCUGAAGGGAAUGAGGAGGAGGAUUCCCACCACGAAAUGGCUGCUAUUCCUUCCGCCCAUUCC--GUUCAGGAACCAAAGCUUUUCGAUCUGGGACUUUUAGGAAGGAAUUGCGCG------------- ..((.(((....)))..(((((((((.............((((((.((((((.((.((((((....(((((.....))).))....))).))).))))))))--.))))))..(((((((((........))).))))..)).))))))))))).------------- ( -48.90, z-score = -0.95, R) >droSec1.super_4 5612108 153 + 6179234 CGGCCUAGUAGUCUAAUGCAGUUCCUACAAAAUUGCGCCUCUGAAGGGAAUGAGGAGGAGGAUUCCCACCACGAAAUGGCUGCUAAUCCUUCCGCCCAUUCC--GUUCAGGAACCAAAGCUUUUCGAUCUGGGACUUUUAGGAAGGAAUUGCGCG------------- ..((.(((....)))..(((((((((.............((((((.((((((.((.((((((((..(((((.....))).))..))))).))).))))))))--.))))))..(((((((((........))).))))..)).))))))))))).------------- ( -51.60, z-score = -1.68, R) >droYak2.chr3R 27714254 153 + 28832112 CGACCUAGCAGCCUAAUGCAGUUCCUCGAAACUUGCGUCUCCGAAGGGAAUGAAAGGGAGGAUUCAGAUCUCGAAAUGGAUACAAAUUCUUGCGCUCAUUCU--GCUCGGGAACCCAAGCUUUUCGAUCUGGGACUUUUGGGCAGUAAUUGCGCG------------- .......((.((((((...((((((((((((((((.((..((((..(((((((..(.(((((((...((((......))))...))))))).)..)))))))--..))))..)).))))..))))))...)))))).)))))).((....)))).------------- ( -54.80, z-score = -2.53, R) >droEre2.scaffold_4820 9268760 153 - 10470090 CGACCUAGUAGCCUAAUGCAGUUCCUCGAAAACUGCGUCUCUGAAGGGAAUGGAAAGGAGGAUGAGGAUCUCGAAAUGGAUACACAUCCUUCCGCCCAUCCU--GUUCAGGAACCCAAGCUUUUCGAUCUGGGACUUUUAGGAAGAAAUUGCCCG------------- ...........(((((...(((((((((((((((..((..(((((.((.((((...((((((((.(.((((......)))).).))))))))...)))).))--.)))))..))...)).)))))))...)))))).))))).............------------- ( -49.90, z-score = -1.72, R) >droAna3.scaffold_12911 1208923 147 - 5364042 CGUCCCAGUAGCUUGAUGCAGUUCCUCAAUCGUUGCGGAAGUGAACAGGACGAGGAGAUGGA--AGUGCCACAGAAUGGC----ACUCCUUGUGCUCGUGCU--GCACAAUCCCGCAAGAUUUUCGAACUGGGACCCGGGGGUACGAGUGUCGUA------------- .((((((((.((.....))...((...((((.((((((...((..(((.(((((.(.(.(((--.((((((.....))))----))))).).).))))).))--)..))...))))))))))...)))))))))).......((((.....))))------------- ( -58.90, z-score = -2.51, R) >dp4.chr2 30417707 168 - 30794189 CGCCCCAGCAGCUUGAUGCAGUUUCUGAAGCUUGGCGACGAGGAUGAGAAGCUGAGCAAUGAUGGGGACAAGGAGAAUACUCCUUCUGCUCCUUCUCCUCCUCAGUCGCAAAGCUGCAGCGACAGCAUGAAGGUGUUUGAGGUGGGAACACAAAAAGCUGCGGAAUUC ...((((((..(((.((((.(((.(((.(((((.((((((((((.((((((..(((((....((....))(((((....)))))..))))))))))).))))).))))).))))).))).))).)))).)))((((((.......)))))).....)))).))..... ( -76.00, z-score = -4.58, R) >droPer1.super_7 1284675 168 - 4445127 CGCCCCAGCAGCUUGAUGCAGUUUCUGAAGCUUGGCGACGAGGAUGAGAAGCUGAGCAAUGAUGGGGACAAGGAGAAUGCUCCUUCUGCUCCUUCUCCUCCUCAGUCGCAAAGCUGCAGCGACAGCAUGAAGGUGUUUGAGGUGGGAACACAAAAAGCUGCGGAAUUC ...((((((..(((.((((.(((.(((.(((((.((((((((((.((((((..(((((....((....))(((((....)))))..))))))))))).))))).))))).))))).))).))).)))).)))((((((.......)))))).....)))).))..... ( -76.20, z-score = -4.17, R) >droWil1.scaffold_181130 10386413 138 + 16660200 CGUCCUAGCAGCGUAAGGCUGUUUCUGCAACUAUGCGGUGAGUUAAAGAGAGAAAAGGAUAGCUCUCAAACUCAGUUGCCUAGUCACAGCAAUAGCCAUAGG--UUUUUUCAGCUAGGAUGUCUUGGCCCUGCUGAUUAC---------------------------- ((((((((((((.((.(((((((.(((..((((.((((((((((...(((((..........))))).)))))).)))).))))..))).))))))).)).)--))......)))))))))..(..((...))..)....---------------------------- ( -50.10, z-score = -3.03, R) >droVir3.scaffold_12855 8564047 139 - 10161210 ----CAGGCAGAGCGAAA-GAUCCAGCCUGCAACUGGGUGACACUAACAUGGAGGAGGACAGGGCUCAGACCCAGGAGCCUUCUCCAUGUCAGUGUACCAGUCAGAGCAAUUACUUUGA--GCUGGGCCCGGAUGCUCGGCCCUAC---------------------- ----.(((..((.(....-).))..(((.(((.((((((.(((((.((((((((((((.(.(((......))).)...)))))))))))).))))).((((((((((......))))))--.)))))))))).)))..))))))..---------------------- ( -66.50, z-score = -4.65, R) >consensus CGGCCUAGCAGCCUAAUGCAGUUCCUCCAAAAUUGCGCCUCUGAAGAGAAUGAGAAGGAGGAUUCGCACAACGAAAUGGCUACUAAUCCUUCCGCCCAUUCU__GUUCAGGAACCCAAGCUUUUCGAUCUGGGACUUUUAGGAAGGAAUUGCGCG_____________ .......(((......)))..................................................................................................................................................... ( -0.63 = -0.83 + 0.20)
| Location | 26,787,049 – 26,787,202 |
|---|---|
| Length | 153 |
| Sequences | 10 |
| Columns | 168 |
| Reading direction | reverse |
| Mean pairwise identity | 51.97 |
| Shannon entropy | 1.00040 |
| G+C content | 0.51591 |
| Mean single sequence MFE | -56.56 |
| Consensus MFE | 0.00 |
| Energy contribution | 0.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 0.00 |
| Mean z-score | -2.83 |
| Structure conservation index | -0.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.833677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26787049 153 - 27905053 -------------CGCGCAAUUCCUUCCUAAGAGUCCCAGAUCGAAAAGCUUGGGUUCCUGAAC--GGAAUGGGCGGAAGGAUUAGCAGCCAUUUCGUUGUGAGAAUCCUCCUCCUCAUUCCCUUCAGAGGGGCAAUUUUGUAGGAACUGCAUUAGGCUACUAGGCCG -------------...(((.(((((.(..((..(((((.((((.((....)).)))).(((((.--((((((((.(((.(((((.(((((......)))))...)))))))).)).)))))).))))).)))))..))..).))))).)))....((((....)))). ( -60.10, z-score = -3.12, R) >droSim1.chr3R 26431359 153 - 27517382 -------------CGCGCAAUUCCUUCCUAAAAGUCCCAGAUCGAAAAGCUUUGGUUCCUGAAC--GGAAUGGGCGGAAGGAAUAGCAGCCAUUUCGUGGUGGGAAUCCUCCUCCUCAUUCCCUUCAGAGGCGCAAUUUUGUAGGAACUGCAUUAGACUACUAGGCCG -------------.(((((.(((((...(((((((.((.(((((((....))))))).(((((.--((((((((.(((.(((......(((((...))))).....)))))).)).)))))).))))).)).))..))))).))))).))).((((....)))))).. ( -47.60, z-score = -0.68, R) >droSec1.super_4 5612108 153 - 6179234 -------------CGCGCAAUUCCUUCCUAAAAGUCCCAGAUCGAAAAGCUUUGGUUCCUGAAC--GGAAUGGGCGGAAGGAUUAGCAGCCAUUUCGUGGUGGGAAUCCUCCUCCUCAUUCCCUUCAGAGGCGCAAUUUUGUAGGAACUGCAUUAGACUACUAGGCCG -------------.(((((.(((((...(((((((.((.(((((((....))))))).(((((.--((((((((.(((.(((((....(((((...)))))...)))))))).)).)))))).))))).)).))..))))).))))).))).((((....)))))).. ( -51.10, z-score = -1.55, R) >droYak2.chr3R 27714254 153 - 28832112 -------------CGCGCAAUUACUGCCCAAAAGUCCCAGAUCGAAAAGCUUGGGUUCCCGAGC--AGAAUGAGCGCAAGAAUUUGUAUCCAUUUCGAGAUCUGAAUCCUCCCUUUCAUUCCCUUCGGAGACGCAAGUUUCGAGGAACUGCAUUAGGCUGCUAGGUCG -------------.(((((.....))).....(((.((...(((((..(((((.((..(((((.--.(((((((.(..((.((((..(((........)))..)))).))..).)))))))..)))))..)).)))))))))))).)))))....((((....)))). ( -42.70, z-score = -0.48, R) >droEre2.scaffold_4820 9268760 153 + 10470090 -------------CGGGCAAUUUCUUCCUAAAAGUCCCAGAUCGAAAAGCUUGGGUUCCUGAAC--AGGAUGGGCGGAAGGAUGUGUAUCCAUUUCGAGAUCCUCAUCCUCCUUUCCAUUCCCUUCAGAGACGCAGUUUUCGAGGAACUGCAUUAGGCUACUAGGUCG -------------.((((...............))))..((((....(((((((((..(((((.--.(((((((.((.((((((.(.(((........))).).)))))).)).)))))))..)))))..))(((((((.....))))))).)))))))....)))). ( -52.96, z-score = -2.16, R) >droAna3.scaffold_12911 1208923 147 + 5364042 -------------UACGACACUCGUACCCCCGGGUCCCAGUUCGAAAAUCUUGCGGGAUUGUGC--AGCACGAGCACAAGGAGU----GCCAUUCUGUGGCACU--UCCAUCUCCUCGUCCUGUUCACUUCCGCAACGAUUGAGGAACUGCAUCAAGCUACUGGGACG -------------((((.....)))).......((((((((((...((((((((((((.((.((--((.(((((.....(((((----(((((...))))))))--))......))))).)))).)).)))))))).))))...)))))((.....))....))))). ( -61.10, z-score = -4.70, R) >dp4.chr2 30417707 168 + 30794189 GAAUUCCGCAGCUUUUUGUGUUCCCACCUCAAACACCUUCAUGCUGUCGCUGCAGCUUUGCGACUGAGGAGGAGAAGGAGCAGAAGGAGUAUUCUCCUUGUCCCCAUCAUUGCUCAGCUUCUCAUCCUCGUCGCCAAGCUUCAGAAACUGCAUCAAGCUGCUGGGGCG ...(((((((((((...(((((.........)))))....((((.((..(((.(((((.(((((.(((((.((((((((((((((((((....))))))..........))))))..)))))).)))))))))).))))).)))..)).)))).))))))).)))).. ( -70.60, z-score = -4.59, R) >droPer1.super_7 1284675 168 + 4445127 GAAUUCCGCAGCUUUUUGUGUUCCCACCUCAAACACCUUCAUGCUGUCGCUGCAGCUUUGCGACUGAGGAGGAGAAGGAGCAGAAGGAGCAUUCUCCUUGUCCCCAUCAUUGCUCAGCUUCUCAUCCUCGUCGCCAAGCUUCAGAAACUGCAUCAAGCUGCUGGGGCG ...(((((((((((...(((((.........)))))....((((.((..(((.(((((.(((((.(((((.((((((((((((((((((....))))))..........))))))..)))))).)))))))))).))))).)))..)).)))).))))))).)))).. ( -69.60, z-score = -4.06, R) >droWil1.scaffold_181130 10386413 138 - 16660200 ----------------------------GUAAUCAGCAGGGCCAAGACAUCCUAGCUGAAAAAA--CCUAUGGCUAUUGCUGUGACUAGGCAACUGAGUUUGAGAGCUAUCCUUUUCUCUCUUUAACUCACCGCAUAGUUGCAGAAACAGCCUUACGCUGCUAGGACG ----------------------------(((((((((((((........)))).))))).....--.....((((.((.(((..((((.((...((((((.(((((..(....)..)))))...))))))..)).))))..))).)).))))))))............ ( -44.20, z-score = -2.62, R) >droVir3.scaffold_12855 8564047 139 + 10161210 ----------------------GUAGGGCCGAGCAUCCGGGCCCAGC--UCAAAGUAAUUGCUCUGACUGGUACACUGACAUGGAGAAGGCUCCUGGGUCUGAGCCCUGUCCUCCUCCAUGUUAGUGUCACCCAGUUGCAGGCUGGAUC-UUUCGCUCUGCCUG---- ----------------------...((((.((((....((..(((((--(.........(((...((((((.((((((((((((((.((((....((((....)))).).))).))))))))))))))...)))))))))))))))..)-)...)))).)))).---- ( -65.60, z-score = -4.33, R) >consensus _____________CGCGCAAUUCCUUCCUAAAAGUCCCAGAUCGAAAAGCUUGGGUUCCUGAAC__AGAAUGAGCGGAAGGAUUAGCAGCCAUUUCGUGGUGCGAAUCCUCCUCCUCAUUCCCUUCAGAGCCGCAAUGUUGGAGGAACUGCAUUAGGCUACUAGGCCG ........................................................................................................................................................................ ( 0.00 = 0.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:41 2011