
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,758,769 – 26,758,859 |
| Length | 90 |
| Max. P | 0.866620 |

| Location | 26,758,769 – 26,758,859 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Shannon entropy | 0.37614 |
| G+C content | 0.47999 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -23.15 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26758769 90 + 27905053 -------------------AAGAGAGGCCGGGGCUCAACGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAAGCGGGC----- -------------------.......(((.((((.....))))...((((((.(((((((((((.(((((......)))))......))))))).)))))))))).)))----- ( -29.10, z-score = -1.34, R) >droSim1.chr3R 26401823 90 + 27517382 -------------------AAGAGAGGCCGGGGCUCAACGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAAGCGGGC----- -------------------.......(((.((((.....))))...((((((.(((((((((((.(((((......)))))......))))))).)))))))))).)))----- ( -29.10, z-score = -1.34, R) >droSec1.super_4 5584791 89 + 6179234 -------------------AAGAGAGGCCGGG-CUCAACGCUUAUGCCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAAGCGGCC----- -------------------......(((((((-(...........)))((((((((((((((((.(((((......)))))......))))))).))))))))))))))----- ( -33.00, z-score = -2.24, R) >droYak2.chr3R 27684472 97 + 28832112 ------------AAGGAAAGAGGGGGUGGGGGGCUCAAGGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAAGCGGGC----- ------------.....(((...((((.....))))....)))...((((((.(((((((((((.(((((......)))))......))))))).))))))))))....----- ( -26.70, z-score = -1.46, R) >droEre2.scaffold_4820 9240634 89 - 10470090 --------------------AAGAGAGGCGGGUCUCCGGGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAAGCGGGC----- --------------------....(((.(.((...)).).)))...((((((.(((((((((((.(((((......)))))......))))))).))))))))))....----- ( -27.90, z-score = -1.20, R) >droAna3.scaffold_12911 1175721 91 - 5364042 ------------------AGAAGAAGCUGAAGGCUCAACGCUUAUGGCUUUUUAAUGGUGCCGUGUGUUGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAAGCAGGC----- ------------------....(((((((.((((.....)))).)))))))(((((((((((((.((((.(((....))).))))..))))))).))))))........----- ( -26.50, z-score = -1.09, R) >dp4.chr2 30387852 114 - 30794189 AGGAGCAAGGGCAUGGCCAGGACAAGCUGAAGGCUCAACGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAACCCUGCCUGCC .........((((.(((.(((..((((((.((((.....)))).))))))((((((((((((((.(((((......)))))......))))))).)))))))..)))))))))) ( -41.70, z-score = -1.34, R) >droPer1.super_7 1254532 114 - 4445127 AGGAGCAAGGGCAUGGCCAGGACAAGCUGAAGGCUCAACGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAACCCUGCCUGCC .........((((.(((.(((..((((((.((((.....)))).))))))((((((((((((((.(((((......)))))......))))))).)))))))..)))))))))) ( -41.70, z-score = -1.34, R) >droWil1.scaffold_181130 10352709 84 + 16660200 -------------------------GCUUCAGGCUCAACGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAACCAUGU----- -------------------------(((..((((.....))))..))).(((((((((((((((.(((((......)))))......))))))).))))))))......----- ( -26.40, z-score = -1.91, R) >droMoj3.scaffold_6540 3992158 84 - 34148556 -------------------------GCUUGAGGCUCAGCGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAAGCAUGU----- -------------------------(((((((((.((.......)))))))(((((((((((((.(((((......)))))......))))))).))))))))))....----- ( -26.20, z-score = -1.12, R) >droVir3.scaffold_13047 11612686 84 + 19223366 -------------------------GCUCGAGGCUCAGCGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAAGCAUGU----- -------------------------(((..((((.....))))..)))((((((((((((((((.(((((......)))))......))))))).))))))))).....----- ( -26.40, z-score = -1.19, R) >droGri2.scaffold_14624 1674352 84 + 4233967 -------------------------GCUUGAGGCUCAAAGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACACUAAAAGCAUGU----- -------------------------((((..((((((((((.....)))).....))).)))((((((((.((((((.....)))))))))))))).....))))....----- ( -26.20, z-score = -1.63, R) >consensus ____________________A___AGCUCGAGGCUCAACGCUUAUGGCUUUUUAAUGGUGCCGUGUGUCGGUUUUGUGGCAAAUAAAAUGGCGCACAUUAAAAGCAGGC_____ .........................(((..((((.....))))..))).(((((((((((((((.(((((......)))))......))))))).))))))))........... (-23.15 = -23.28 + 0.13)

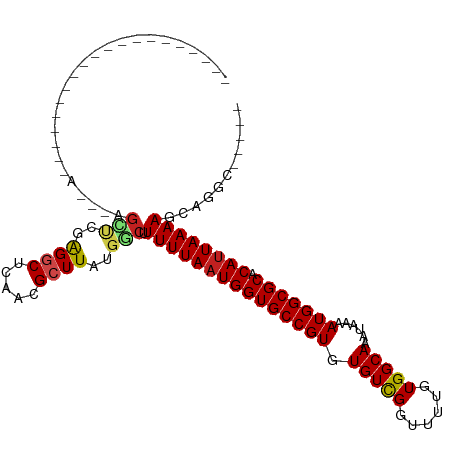
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:36 2011