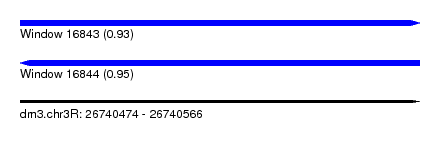
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,740,474 – 26,740,566 |
| Length | 92 |
| Max. P | 0.950889 |

| Location | 26,740,474 – 26,740,566 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Shannon entropy | 0.41836 |
| G+C content | 0.38807 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.46 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
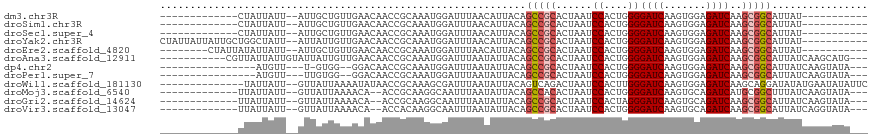
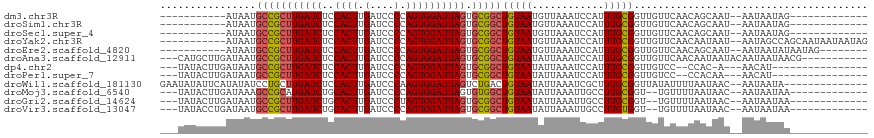
>dm3.chr3R 26740474 92 + 27905053 -------------CUAUUAUU--AUUGCUGUUGAACAACCGCAAAUGGAUUUAACAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGGAGAUCAAGCGGCAUUAU----------- -------------........--..(((.(((....))).)))((((.......))))...(((((.....((((((........))))))......))))).....----------- ( -25.00, z-score = -2.54, R) >droSim1.chr3R 26383621 92 + 27517382 -------------CUAUUAUU--AUUGCUGUUGAACAACCGCAAAUGGAUUUAACAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGGAGAUCAAGCGGCAUUAU----------- -------------........--..(((.(((....))).)))((((.......))))...(((((.....((((((........))))))......))))).....----------- ( -25.00, z-score = -2.54, R) >droSec1.super_4 5566630 92 + 6179234 -------------CUAUUAUU--AUUGCUGUUGAACAACCGCAAAUGGAUUUAACAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGGAGAUCAAGCGGCAUUAU----------- -------------........--..(((.(((....))).)))((((.......))))...(((((.....((((((........))))))......))))).....----------- ( -25.00, z-score = -2.54, R) >droYak2.chr3R 27665557 105 + 28832112 CUAUUAUUAUUGCUGGCUAUU--AUUAUUGUUGAACAACCGCAAAUGGAUUUAACAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGGAGAUCAAGCGGCAUUAU----------- .....................--.....(((((((...(((....))).))))))).....(((((.....((((((........))))))......))))).....----------- ( -24.10, z-score = -1.15, R) >droEre2.scaffold_4820 9222342 97 - 10470090 --------CUAUUAUAUUAUU--AUUGCUGUUGAACAACCGCAAAUGGAUUUAACAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGGAGAUCAAGCGGCAUUAU----------- --------.............--..(((.(((....))).)))((((.......))))...(((((.....((((((........))))))......))))).....----------- ( -25.00, z-score = -2.41, R) >droAna3.scaffold_12911 1156559 104 - 5364042 -----------CGUUAUUAUUGUAUUAUUGUUGAACAACCGCAAAUGGAUUUAAUAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGGAGAUCAAGCGGCAUUAUCAAGCAUG--- -----------.(((.....((((....(((((((...(((....))).))))))).))))(((((.....((((((........))))))......))))).......)))...--- ( -25.70, z-score = -1.72, R) >dp4.chr2 30367923 93 - 30794189 ----------------AUGUU---U-GUGG--GGACAACCGCAAAUGGAUUUAAUAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGGAGAUCAAGCGGCAUUAUCAAGUAUA--- ----------------..(((---(-((((--......))))))))...............(((((.....((((((........))))))......))))).............--- ( -29.50, z-score = -3.09, R) >droPer1.super_7 1234359 94 - 4445127 ----------------AUGUU---UUGUGG--GGACAACCGCAAAUGGAUUUAAUAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGGAGAUCAAGCGGCAUUAUCAAGUAUA--- ----------------....(---((((((--......)))))))................(((((.....((((((........))))))......))))).............--- ( -28.00, z-score = -2.57, R) >droWil1.scaffold_181130 10330123 102 + 16660200 --------------UAUUAUU--GUUAUUAAAAUAUAACCGCAAAGCGAUUUAAUAUUACAGUCAGACUAAUCCACUUGGGAUCAAGUGGAGAUCAAGCAGGAUAUAUGAAUAUAUUC --------------.......--..((((...(((((.((((.....((((.........)))).((....((((((((....))))))))..))..)).)).))))).))))..... ( -19.80, z-score = -1.76, R) >droMoj3.scaffold_6540 3969556 98 - 34148556 -------------UUAUUAUU--GUUAUUAAAACA--ACCGCAAGGCAAUUUAAUAUUACAGCCACACUAAUCCACUGGGGAUCAAGUGCAGAUCAUGCGGCUUUAUCAAGUAUA--- -------------......((--(((.....))))--)(((((.(((..............))).((((.((((.....))))..)))).......)))))..............--- ( -18.94, z-score = -1.12, R) >droGri2.scaffold_14624 1643538 98 + 4233967 -------------UUAUUAUU--GUUAUUAAAACA--ACCGCAAGGCAAUUUAAUAUUACAGCCGCACUAAUCCACUAGGGAUCAAGUGCAGAUCAAGCGGCAUUAUCAAGUAUA--- -------------......((--(((.....))))--)((((..(((..............)))(((((.((((.....))))..))))).......))))..............--- ( -22.64, z-score = -3.11, R) >droVir3.scaffold_13047 11589644 98 + 19223366 -------------UUAUUAUU--GUUAUUAAAACA--ACCACAAGGCAAUUUAAUAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGCAGAUCAAGCGGCAUUAUCAGGUAUA--- -------------.......(--(((((((((...--.((....))...)))))))..)))(((((......((....))((((.......))))..))))).............--- ( -18.60, z-score = -1.16, R) >consensus _____________UUAUUAUU__AUUAUUGUUGAACAACCGCAAAUGGAUUUAAUAUUACAGCCGCACUAAUCCACUGGGGAUCAAGUGGAGAUCAAGCGGCAUUAUCAAGUAUA___ .............................................................(((((......((....))((((.......))))..)))))................ (-13.12 = -13.46 + 0.34)
| Location | 26,740,474 – 26,740,566 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Shannon entropy | 0.41836 |
| G+C content | 0.38807 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -14.95 |
| Energy contribution | -14.99 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
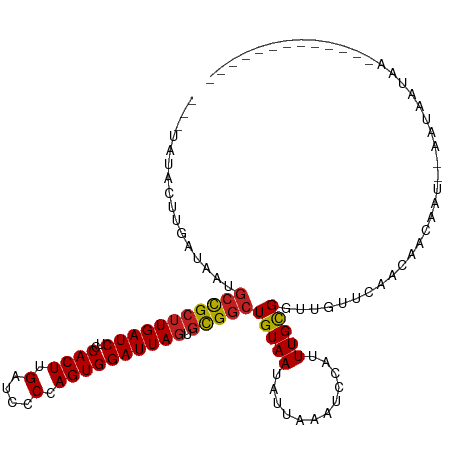
>dm3.chr3R 26740474 92 - 27905053 -----------AUAAUGCCGCUUGAUCUCCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUGUUAAAUCCAUUUGCGGUUGUUCAACAGCAAU--AAUAAUAG------------- -----------.....(((((((((..((((((.(....).)))))))))).))))).................((((.(((....))).)))).--........------------- ( -23.00, z-score = -1.71, R) >droSim1.chr3R 26383621 92 - 27517382 -----------AUAAUGCCGCUUGAUCUCCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUGUUAAAUCCAUUUGCGGUUGUUCAACAGCAAU--AAUAAUAG------------- -----------.....(((((((((..((((((.(....).)))))))))).))))).................((((.(((....))).)))).--........------------- ( -23.00, z-score = -1.71, R) >droSec1.super_4 5566630 92 - 6179234 -----------AUAAUGCCGCUUGAUCUCCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUGUUAAAUCCAUUUGCGGUUGUUCAACAGCAAU--AAUAAUAG------------- -----------.....(((((((((..((((((.(....).)))))))))).))))).................((((.(((....))).)))).--........------------- ( -23.00, z-score = -1.71, R) >droYak2.chr3R 27665557 105 - 28832112 -----------AUAAUGCCGCUUGAUCUCCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUGUUAAAUCCAUUUGCGGUUGUUCAACAAUAAU--AAUAGCCAGCAAUAAUAAUAG -----------.....(((((((((..((((((.(....).)))))))))).))))).................(((((((((((..........--))))))).))))......... ( -26.50, z-score = -2.19, R) >droEre2.scaffold_4820 9222342 97 + 10470090 -----------AUAAUGCCGCUUGAUCUCCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUGUUAAAUCCAUUUGCGGUUGUUCAACAGCAAU--AAUAAUAUAAUAG-------- -----------.....(((((((((..((((((.(....).)))))))))).)))))(((.((((((.......((((.(((....))).)))).--..)))))).))).-------- ( -24.30, z-score = -2.06, R) >droAna3.scaffold_12911 1156559 104 + 5364042 ---CAUGCUUGAUAAUGCCGCUUGAUCUCCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUAUUAAAUCCAUUUGCGGUUGUUCAACAAUAAUACAAUAAUAACG----------- ---..((.(((((((((((((((((..((((((.(....).)))))))))).)))))(((((............))))))))).)))))).................----------- ( -22.00, z-score = -0.97, R) >dp4.chr2 30367923 93 + 30794189 ---UAUACUUGAUAAUGCCGCUUGAUCUCCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUAUUAAAUCCAUUUGCGGUUGUCC--CCAC-A---AACAU---------------- ---.....((((((..(((((((((..((((((.(....).)))))))))).))))).....)))))).....((((.((......--)).)-)---))...---------------- ( -22.60, z-score = -1.90, R) >droPer1.super_7 1234359 94 + 4445127 ---UAUACUUGAUAAUGCCGCUUGAUCUCCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUAUUAAAUCCAUUUGCGGUUGUCC--CCACAA---AACAU---------------- ---.....((((((..(((((((((..((((((.(....).)))))))))).))))).....)))))).....((((.((......--)).)))---)....---------------- ( -22.90, z-score = -2.06, R) >droWil1.scaffold_181130 10330123 102 - 16660200 GAAUAUAUUCAUAUAUCCUGCUUGAUCUCCACUUGAUCCCAAGUGGAUUAGUCUGACUGUAAUAUUAAAUCGCUUUGCGGUUAUAUUUUAAUAAC--AAUAAUA-------------- ((((((.................(((.((((((((....))))))))...))).((((((((............)))))))))))))).......--.......-------------- ( -19.20, z-score = -1.74, R) >droMoj3.scaffold_6540 3969556 98 + 34148556 ---UAUACUUGAUAAAGCCGCAUGAUCUGCACUUGAUCCCCAGUGGAUUAGUGUGGCUGUAAUAUUAAAUUGCCUUGCGGU--UGUUUUAAUAAC--AAUAAUAA------------- ---.....((((...(((((((.(..(..((((.(((((.....)))))))))..)..(((((.....)))))).))))))--)...))))....--........------------- ( -24.80, z-score = -2.21, R) >droGri2.scaffold_14624 1643538 98 - 4233967 ---UAUACUUGAUAAUGCCGCUUGAUCUGCACUUGAUCCCUAGUGGAUUAGUGCGGCUGUAAUAUUAAAUUGCCUUGCGGU--UGUUUUAAUAAC--AAUAAUAA------------- ---.....((((....(((((..(..(((((((.(((((.....))))))))))))..(((((.....))))))..)))))--....))))....--........------------- ( -25.40, z-score = -2.69, R) >droVir3.scaffold_13047 11589644 98 - 19223366 ---UAUACCUGAUAAUGCCGCUUGAUCUGCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUAUUAAAUUGCCUUGUGGU--UGUUUUAAUAAC--AAUAAUAA------------- ---.............(((((((((((..(............)..)))))).)))))(((..(((((((..(((....)))--...)))))))))--).......------------- ( -24.80, z-score = -2.48, R) >consensus ___UAUACUUGAUAAUGCCGCUUGAUCUCCACUUGAUCCCCAGUGGAUUAGUGCGGCUGUAAUAUUAAAUCCAUUUGCGGUUGUUCAACAACAAU__AAUAAUAA_____________ ................(((((((((((..((((.(....).)))))))))).)))))(((((............)))))....................................... (-14.95 = -14.99 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:35 2011