
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,729,483 – 26,729,578 |
| Length | 95 |
| Max. P | 0.500000 |

| Location | 26,729,483 – 26,729,578 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 67.32 |
| Shannon entropy | 0.61788 |
| G+C content | 0.55692 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -14.26 |
| Energy contribution | -16.08 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 26729483 95 + 27905053 --AAUGGACCGCAAGUGGAAGAAGACCCAAU--CCGCUUCUGUGGCAGGUAGCAUCCGA--GUCCGGCCCGGGAUCCUGUUACUG--UUCCUUCAGCGAGUCC------ --...((((((((((((((...........)--))))))..((((((((.....((((.--(.....).))))..))))))))..--........))).))))------ ( -31.40, z-score = -0.79, R) >droWil1.scaffold_181130 10316489 95 + 16660200 --CGAGCAAAGCAAGUGAAAGUGGCUUUGACAGUGGCUUUUGGGUCAGGUAGCAUUUUGCUCUUUUGUGCCCAGCCAUGUCGAAAUUCUUCUGGUUU------------ --.(((((((((...(....)..)))))((((.(((((...((((((((.(((.....)))..)))).)))))))))))))............))))------------ ( -26.20, z-score = 0.01, R) >droSim1.chr3R 26372649 95 + 27517382 --AACGGACCGCAAGUGGAAGAAGACCCAAU--CCGCUUCUGUGGCAGGUAGCAUCCGA--GUCCGGUCCGGGAUCCUGUUACUG--UUCCUUCUGCGAGUCC------ --...((((((((...((((((((.......--...)))))((((((((...(..(((.--...))).....)..))))))))..--.)))...)))).))))------ ( -32.10, z-score = -0.85, R) >droSec1.super_4 5555703 95 + 6179234 --AACGGACCGCAAGUGGAAGAAGACCCAAU--CCGCUUCUGUGGCAGGUAGCUUCCGA--GUCCGGUCCGGGAUCCUGUUACUG--UUCCUCCUGCGAGUCC------ --...((((((((.(.((((((((.......--...)))))((((((((....(((((.--(......)))))).))))))))..--.))).).)))).))))------ ( -34.90, z-score = -1.54, R) >droYak2.chr3R 27654044 95 + 28832112 --AAUGGACCGCAAGUGGAGGAGAACCCCAU--CCGCUUCCGUGGCAGGUAGCCUUCGA--GUCCGGUCCGGAGUCCUGUUACUG--UUCCUCCUGCGAGUCC------ --...((((((((...(((((((........--((((....))))..((((((....((--.((((...)))).))..)))))).--))))))))))).))))------ ( -36.50, z-score = -1.81, R) >droEre2.scaffold_4820 9211101 97 - 10470090 UAUGUGGACCGCAAUUGGAAGAAGACCCAAU--CCGCUUGUGUGGCAGGUAGUAUUCAA--GUCCGGGCCGGAAUCCUGUUACUG--UUCUUUCUGCGAGUCC------ .....((((((((...(((((((.(((....--((((....))))..)))((((.....--.((((...)))).......)))).--))))))))))).))))------ ( -28.82, z-score = -0.72, R) >droAna3.scaffold_12911 1142205 105 - 5364042 --AGCGGGCCACAAGUGGCAGAAAACUCAAU--CCGCAUUUGUGGCAGGUAGCAUCCGGAGGACUGAGUCCGGUUGUAACUACGGCCUUCGCACUCUGGAUCUCGGUUC --.((..((((((((((.(.((........)--).))))))))))).....))((((((((..(.(((.(((..........))).))).)..))))))))........ ( -37.20, z-score = -1.23, R) >consensus __AACGGACCGCAAGUGGAAGAAGACCCAAU__CCGCUUCUGUGGCAGGUAGCAUCCGA__GUCCGGUCCGGGAUCCUGUUACUG__UUCCUUCUGCGAGUCC______ .....((((((((...((((((((............)))))((((((((......(((......)))........)))))))).....)))...)))).))))...... (-14.26 = -16.08 + 1.82)

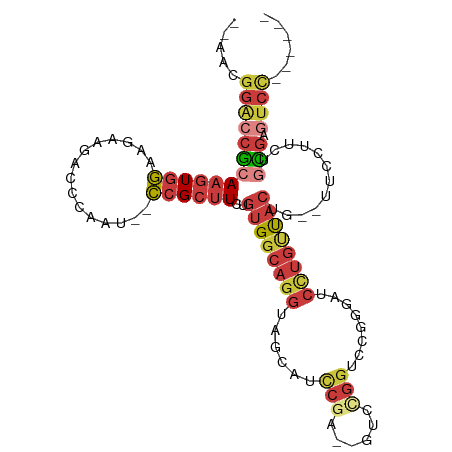
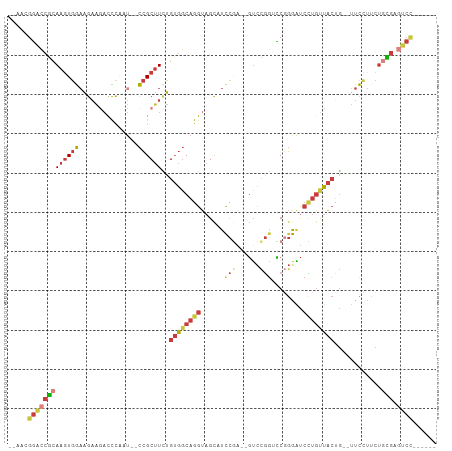
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:32 2011