| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,724,738 – 26,724,797 |
| Length | 59 |
| Max. P | 0.983883 |

| Location | 26,724,738 – 26,724,797 |
|---|---|
| Length | 59 |
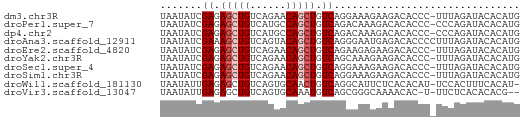
| Sequences | 10 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Shannon entropy | 0.40568 |
| G+C content | 0.43952 |
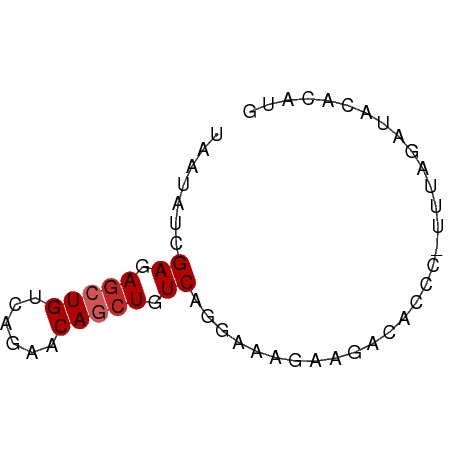
| Mean single sequence MFE | -13.03 |
| Consensus MFE | -9.00 |
| Energy contribution | -9.30 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
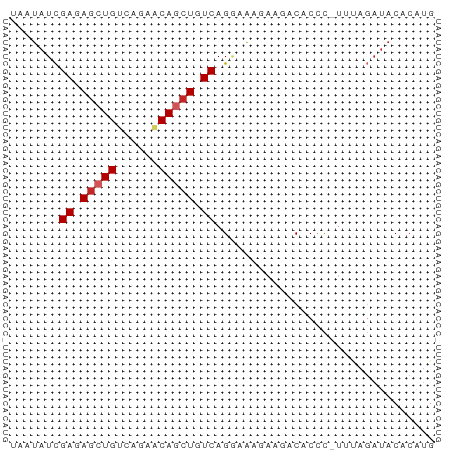
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26724738 59 + 27905053 UAAUAUCGAGAGCUGUCAGAACAGCUGUCAGGAAAGAAGACACCC-UUUAGAUACACAUG ...((((((.((((((....)))))).))......((((.....)-))).))))...... ( -14.40, z-score = -2.76, R) >droPer1.super_7 1216315 59 - 4445127 UAAUAUCGAGAGCUGUCAUGCCAGCUGUCAGACAAAGACACACCC-CCCAGAUACACAUG ...((((...(((((......)))))(((.......)))......-....))))...... ( -10.20, z-score = -1.80, R) >dp4.chr2 30347675 59 - 30794189 UAAUAUCGAGAGCUGUCAUGCCAGCUGUCAGACAAAGACACACCC-CCCAGAUACACAUG ...((((...(((((......)))))(((.......)))......-....))))...... ( -10.20, z-score = -1.80, R) >droAna3.scaffold_12911 1134496 60 - 5364042 UAAUAUCGAAAGCUGUCAGUACAGCUGUCAGGGAAUGAGACACCCCUUUAGAUACACAUG ...((((((.((((((....)))))).))((((..........))))...))))...... ( -15.00, z-score = -2.19, R) >droEre2.scaffold_4820 9204758 59 - 10470090 UAAUAUCGAGAGCUGUCAGAACAGCUGUCAGAAGAGAAGACACCC-UUUAGAUACACAUG ...((((((.((((((....)))))).))......((((.....)-))).))))...... ( -14.40, z-score = -2.97, R) >droYak2.chr3R 27648809 59 + 28832112 UAAUAUCGAGAGCUGUCAGAACAGCUGUCAGCAAAGAAGACACCC-UUUAGAUACACAUG ...((((((.((((((....)))))).))......((((.....)-))).))))...... ( -14.40, z-score = -3.02, R) >droSec1.super_4 5550398 59 + 6179234 UAAUAUCGAGAGCUGUCAGAACAGCUGUCAGGAAAGAAGACACCC-UUUAGAUACACAUG ...((((((.((((((....)))))).))......((((.....)-))).))))...... ( -14.40, z-score = -2.76, R) >droSim1.chr3R 26366338 59 + 27517382 UAAUAUCGAGAGCUGUCAGAACAGCUGUCAGGAAAGAAGACACCC-UUUAGAUACACAUG ...((((((.((((((....)))))).))......((((.....)-))).))))...... ( -14.40, z-score = -2.76, R) >droWil1.scaffold_181130 10312600 58 + 16660200 UAAUAUUGAGAGCUGUCAGUGCAACUGUCAGGCAUUCUCACACAU-UCCACUUUCACAU- ......(((((((((.(((.....))).)))...)))))).....-.............- ( -11.50, z-score = -1.22, R) >droVir3.scaffold_13047 11572272 56 + 19223366 UAAUAUUGAGAGCUGUCAGUGCAAAUGUCAGCGGGCAAAACAC-U-UUCUCACACACG-- ......(((((((((.((.......)).))))((........)-)-.)))))......-- ( -11.40, z-score = -0.86, R) >consensus UAAUAUCGAGAGCUGUCAGAACAGCUGUCAGGAAAGAAGACACCC_UUUAGAUACACAUG .......((.(((((......))))).))............................... ( -9.00 = -9.30 + 0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:29 2011