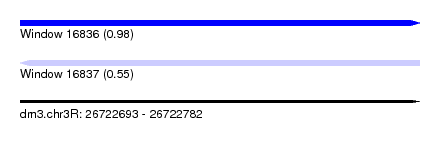
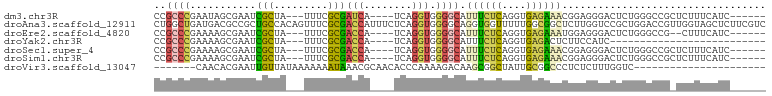
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,722,693 – 26,722,782 |
| Length | 89 |
| Max. P | 0.979564 |

| Location | 26,722,693 – 26,722,782 |
|---|---|
| Length | 89 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 57.03 |
| Shannon entropy | 0.79899 |
| G+C content | 0.55062 |
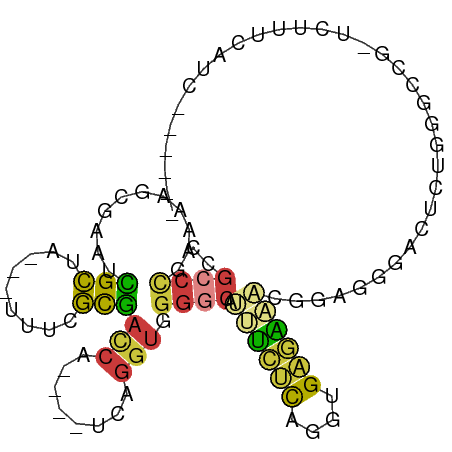
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.46 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.70 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
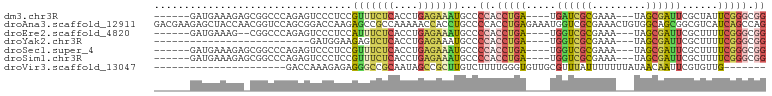
>dm3.chr3R 26722693 89 + 27905053 ------GAUGAAAGAGCGGCCCAGAGUCCCUCCGUUUCUCACCUGAGAAAUGCCCCACCUGA----UGAUCGCGAAA---UAGCGAUUCGCUAUUCGGGCGG ------.......(((.(((.....))).)))((((((((....))))))))..((.(((((----.((((((....---..))))))......))))).)) ( -28.00, z-score = -1.50, R) >droAna3.scaffold_12911 1132482 102 - 5364042 GACGAAGAGCUACCAACGGUCCAGCGGACCAAGAGCCGCCAAAAACCACCUGCCCCACCUGAGAAAUGGUCGCGAAACUGUGGCAGCGGCGUCAUCAGCCAG (((..............(((((...)))))....(((((......((((.(((...(((........))).))).....))))..))))))))......... ( -27.50, z-score = -0.04, R) >droEre2.scaffold_4820 9202807 87 - 10470090 ------GAUGAAAG--CGGCCCAGAGUCCCUCCAUUUCUCACCUGAGAAAUGCCCCACCUGA----UGGUCGCGAAA---UAGCGAUUCGCUUUUCGGGCGG ------.......(--((((((((.((.....((((((((....))))))))....))))).----.))))))((((---.((((...))))))))...... ( -28.10, z-score = -1.44, R) >droYak2.chr3R 27646886 69 + 28832112 --------------------------GAUGGAAGAGUCUCACCUGAGAAAUGCCCCACCUGA----UGGUCGCGAAA---UAGCGAUUCGCUUUUCGGGCGG --------------------------...((.....((((....))))....))((.(((((----.((((((....---..))))))......))))).)) ( -20.50, z-score = -0.59, R) >droSec1.super_4 5548388 89 + 6179234 ------GAUGAAAGAGCGGCCCAGAGUCCCUCCGUUUCUCACCUGAGAAAUGCCCCACCUGA----UGGUCGCGAAA---UAGCGAUUCGCUUUUCGGGCGG ------.........(((((((((.((.....((((((((....))))))))....))))).----.))))))((((---.((((...))))))))...... ( -27.70, z-score = -0.70, R) >droSim1.chr3R 26364296 89 + 27517382 ------GAUGAAAGAGCGGCCCAGAGUCCCUCCGUUUCUCACCUGAGAAAUGCCCCACCUGA----UGGUCGCGAAA---UAGCGAUUCGCUUUUCGGGCGG ------.........(((((((((.((.....((((((((....))))))))....))))).----.))))))((((---.((((...))))))))...... ( -27.70, z-score = -0.70, R) >droVir3.scaffold_13047 11563192 73 + 19223366 ----------------------GACCAAAGAGAGGGCCGCAAUAGCCGCUUGUCUUUUGGGUGUUGCGUUUAUUUUUUUAUAACAAUUCGUGUUG------- ----------------------....(((((((..(.(((((((.(((.........))).))))))).)..))))))).(((((.....)))))------- ( -15.90, z-score = -0.30, R) >consensus ______GAUGAAAGA_CGGCCCAGAGUCCCUCCGUUUCUCACCUGAGAAAUGCCCCACCUGA____UGGUCGCGAAA___UAGCGAUUCGCUUUUCGGGCGG .................................(((((((....)))))))...((.(((((.....((((((.........))))))......))))).)) (-14.18 = -14.46 + 0.28)
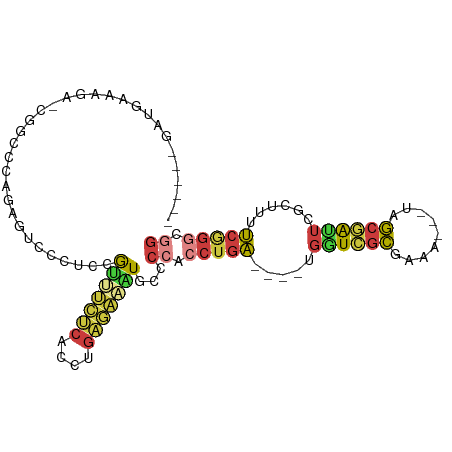
| Location | 26,722,693 – 26,722,782 |
|---|---|
| Length | 89 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 57.03 |
| Shannon entropy | 0.79899 |
| G+C content | 0.55062 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -10.95 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26722693 89 - 27905053 CCGCCCGAAUAGCGAAUCGCUA---UUUCGCGAUCA----UCAGGUGGGGCAUUUCUCAGGUGAGAAACGGAGGGACUCUGGGCCGCUCUUUCAUC------ (((((.((...(((((......---.))))).....----)).)))))(((.((((((....))))))(((((...))))).)))...........------ ( -30.00, z-score = -0.71, R) >droAna3.scaffold_12911 1132482 102 + 5364042 CUGGCUGAUGACGCCGCUGCCACAGUUUCGCGACCAUUUCUCAGGUGGGGCAGGUGGUUUUUGGCGGCUCUUGGUCCGCUGGACCGUUGGUAGCUCUUCGUC ..(((((..((((((((((((((.((..(((.............)))..))..)))))....))))))....(((((...))))))))..)))))....... ( -39.42, z-score = -0.72, R) >droEre2.scaffold_4820 9202807 87 + 10470090 CCGCCCGAAAAGCGAAUCGCUA---UUUCGCGACCA----UCAGGUGGGGCAUUUCUCAGGUGAGAAAUGGAGGGACUCUGGGCCG--CUUUCAUC------ (((((.((...(((((......---.))))).....----)).)))))((((((((((....)))))))((((...))))..))).--........------ ( -28.50, z-score = -0.40, R) >droYak2.chr3R 27646886 69 - 28832112 CCGCCCGAAAAGCGAAUCGCUA---UUUCGCGACCA----UCAGGUGGGGCAUUUCUCAGGUGAGACUCUUCCAUC-------------------------- ..((((.....(((((......---.))))).(((.----...))).))))...((((....))))..........-------------------------- ( -20.60, z-score = -0.97, R) >droSec1.super_4 5548388 89 - 6179234 CCGCCCGAAAAGCGAAUCGCUA---UUUCGCGACCA----UCAGGUGGGGCAUUUCUCAGGUGAGAAACGGAGGGACUCUGGGCCGCUCUUUCAUC------ (((((.((...(((((......---.))))).....----)).)))))(((.((((((....))))))(((((...))))).)))...........------ ( -30.00, z-score = -0.52, R) >droSim1.chr3R 26364296 89 - 27517382 CCGCCCGAAAAGCGAAUCGCUA---UUUCGCGACCA----UCAGGUGGGGCAUUUCUCAGGUGAGAAACGGAGGGACUCUGGGCCGCUCUUUCAUC------ (((((.((...(((((......---.))))).....----)).)))))(((.((((((....))))))(((((...))))).)))...........------ ( -30.00, z-score = -0.52, R) >droVir3.scaffold_13047 11563192 73 - 19223366 -------CAACACGAAUUGUUAUAAAAAAAUAAACGCAACACCCAAAAGACAAGCGGCUAUUGCGGCCCUCUCUUUGGUC---------------------- -------.((((.....)))).....................((((((((...((.((....)).))..))).)))))..---------------------- ( -11.60, z-score = -0.44, R) >consensus CCGCCCGAAAAGCGAAUCGCUA___UUUCGCGACCA____UCAGGUGGGGCAUUUCUCAGGUGAGAAACGGAGGGACUCUGGGCCG_UCUUUCAUC______ (((((.((........((((.........)))).......)).)))))....((((((....)))))).................................. (-10.95 = -11.12 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:29 2011