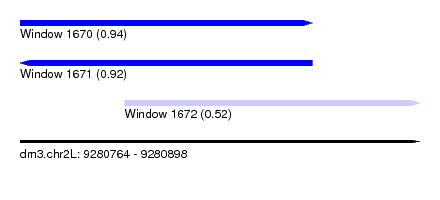
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,280,764 – 9,280,898 |
| Length | 134 |
| Max. P | 0.940113 |

| Location | 9,280,764 – 9,280,862 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.41 |
| Shannon entropy | 0.23252 |
| G+C content | 0.47231 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -20.32 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
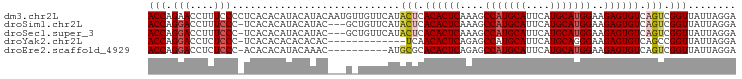
>dm3.chr2L 9280764 98 + 23011544 ACCAGAACCUUUCCCCUCACACAUACAUACAAUGUUGUUCAUACUCACACUCAAAGCCAUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGA .((..((((...........(((.((((...)))))))....(((.((((((....(((((((....)))))))..)))))).))).))))....)). ( -26.40, z-score = -2.93, R) >droSim1.chr2L 9057036 94 + 22036055 ACCAGGACCUUUCCC-UCACACAUACAUAC---GCUGUUCAUACUCACACUCAAAGCCAUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGA .((.(((....))).-..............---((((.....(((.((((((....(((((((....)))))))..)))))).))))))).....)). ( -25.40, z-score = -2.40, R) >droSec1.super_3 4739719 94 + 7220098 ACCAGGACCUUUCCC-UCACACAUACAUAC---GCUGUUCAUACUCACACUCAAAGCCAUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGA .((.(((....))).-..............---((((.....(((.((((((....(((((((....)))))))..)))))).))))))).....)). ( -25.40, z-score = -2.40, R) >droYak2.chr2L 11949539 84 + 22324452 ACCAGGACCUCUCCC-UCACACACACACAC-------------UCAACACUCAGAGCCAUGCAUUCAUGCAGGGAAGAGUGUCAGCCGGUUAUUAGGA (((..((((((((((-.............(-------------((........)))...((((....))))))).)))).)))....)))........ ( -20.10, z-score = -0.67, R) >droEre2.scaffold_4929 9887999 87 + 26641161 ACCAGGACCUCUCCC-ACACACAUACAAAC----------AUGCGCACACUCAGAGCCAUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGA .((.(((....))).-...........(((----------..((..((((((....(((((((....)))))))..))))))..))..)))....)). ( -23.80, z-score = -1.70, R) >consensus ACCAGGACCUUUCCC_UCACACAUACAUAC___G_UGUUCAUACUCACACUCAAAGCCAUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGA (((.(((....)))............................(((.((((((....(((((((....)))))))..)))))).))).)))........ (-20.32 = -21.04 + 0.72)
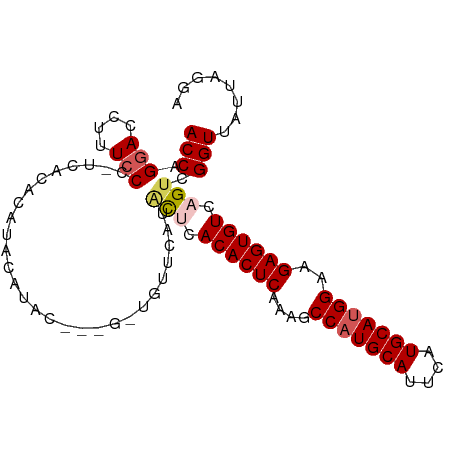
| Location | 9,280,764 – 9,280,862 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 86.41 |
| Shannon entropy | 0.23252 |
| G+C content | 0.47231 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.72 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9280764 98 - 23011544 UCCUAAUAACCGACUGACACUCUUCCAUGCAUGAAUGCAUGGCUUUGAGUGUGAGUAUGAACAACAUUGUAUGUAUGUGUGAGGGGAAAGGUUCUGGU ........(((((((.((((((..(((((((....)))))))....)))))).)))..((((..(.(..(((....)))..)..).....)))))))) ( -29.30, z-score = -1.87, R) >droSim1.chr2L 9057036 94 - 22036055 UCCUAAUAACCGACUGACACUCUUCCAUGCAUGAAUGCAUGGCUUUGAGUGUGAGUAUGAACAGC---GUAUGUAUGUGUGA-GGGAAAGGUCCUGGU ........(((.(((.((((((..(((((((....)))))))....)))))).)))....(((.(---((....))))))..-((((....))))))) ( -28.70, z-score = -1.85, R) >droSec1.super_3 4739719 94 - 7220098 UCCUAAUAACCGACUGACACUCUUCCAUGCAUGAAUGCAUGGCUUUGAGUGUGAGUAUGAACAGC---GUAUGUAUGUGUGA-GGGAAAGGUCCUGGU ........(((.(((.((((((..(((((((....)))))))....)))))).)))....(((.(---((....))))))..-((((....))))))) ( -28.70, z-score = -1.85, R) >droYak2.chr2L 11949539 84 - 22324452 UCCUAAUAACCGGCUGACACUCUUCCCUGCAUGAAUGCAUGGCUCUGAGUGUUGA-------------GUGUGUGUGUGUGA-GGGAGAGGUCCUGGU ........(((((..(((.(((((((..((((..(((((((.(((........))-------------))))))))))))..-))))))))))))))) ( -31.00, z-score = -2.56, R) >droEre2.scaffold_4929 9887999 87 - 26641161 UCCUAAUAACCGACUGACACUCUUCCAUGCAUGAAUGCAUGGCUCUGAGUGUGCGCAU----------GUUUGUAUGUGUGU-GGGAGAGGUCCUGGU ........((((...(((.(((((((((((((....(((((((.(.....).)).)))----------))......))))))-)))))))))).)))) ( -32.50, z-score = -2.82, R) >consensus UCCUAAUAACCGACUGACACUCUUCCAUGCAUGAAUGCAUGGCUUUGAGUGUGAGUAUGAACA_C___GUAUGUAUGUGUGA_GGGAAAGGUCCUGGU ........(((.(((.((((((..(((((((....)))))))....)))))).)))...........................((((....))))))) (-23.00 = -23.72 + 0.72)

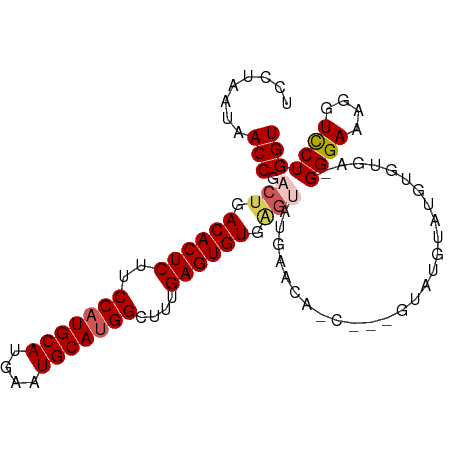
| Location | 9,280,799 – 9,280,898 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.91 |
| Shannon entropy | 0.33277 |
| G+C content | 0.51876 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -20.26 |
| Energy contribution | -21.35 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
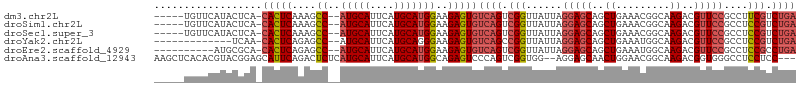
>dm3.chr2L 9280799 99 + 23011544 -----UGUUCAUACUCA-CACUCAAAGCC--AUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGAGCAGCUGAAACGGCAAGACGUUCCGCCUUCGUCUGA -----...........(-(((((....((--(((((....)))))))..))))))(((.(((......(((((.((((...)))).....)))))....))).))). ( -31.40, z-score = -1.91, R) >droSim1.chr2L 9057067 99 + 22036055 -----UGUUCAUACUCA-CACUCAAAGCC--AUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGAGCAGCUGAAACGGCAAGACGUUCCGCCUCCGUCUGA -----...........(-(((((....((--(((((....)))))))..))))))(((.(((......(((((.((((...)))).....)))))....))).))). ( -33.90, z-score = -2.64, R) >droSec1.super_3 4739750 99 + 7220098 -----UGUUCAUACUCA-CACUCAAAGCC--AUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGAGCAGCUGAAACGGCAAGACGUUCCGCCUCCGUCUGA -----...........(-(((((....((--(((((....)))))))..))))))(((.(((......(((((.((((...)))).....)))))....))).))). ( -33.90, z-score = -2.64, R) >droYak2.chr2L 11949568 91 + 22324452 -------------UCAA-CACUCAGAGCC--AUGCAUUCAUGCAGGGAAGAGUGUCAGCCGGUUAUUAGGAGCAGCUGAAAUGGCAAGACGUUCCGCCUCCGUCUGA -------------...(-(((((....((--.((((....)))).))..))))))(((.(((......(((((.(((.....))).....)))))....))).))). ( -27.90, z-score = -0.74, R) >droEre2.scaffold_4929 9888028 94 + 26641161 ----------AUGCGCA-CACUCAGAGCC--AUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGAGCAGCUGAAAUGGCAAGACGUUCCGCCUCCGCCUGA ----------......(-(((((....((--(((((....)))))))..))))))(((.(((......(((((.(((.....))).....)))))....))).))). ( -32.20, z-score = -1.25, R) >droAna3.scaffold_12943 621250 102 + 5039921 AAGCUCACACGUACGGAGCAUUCAGACUCUCAUGCAUUCAUGCAUGGCAGAGUCCCAGUCGGUGG--AGGAGCAACUGGAACGGCAAGACGGUGGGCCUCCUCC--- ..((((.(......))))).....((((((((((((....))))))..))))))((....)).((--((((((.((((...........))))..).)))))))--- ( -35.40, z-score = -0.78, R) >consensus _____UGUUCAUACUCA_CACUCAAAGCC__AUGCAUUCAUGCAUGGAAGAGUGUCAGUCGGUUAUUAGGAGCAGCUGAAACGGCAAGACGUUCCGCCUCCGUCUGA ..................(((((....((..(((((....)))))))..)))))((((.(((......(((((..((.........))..)))))....))).)))) (-20.26 = -21.35 + 1.09)
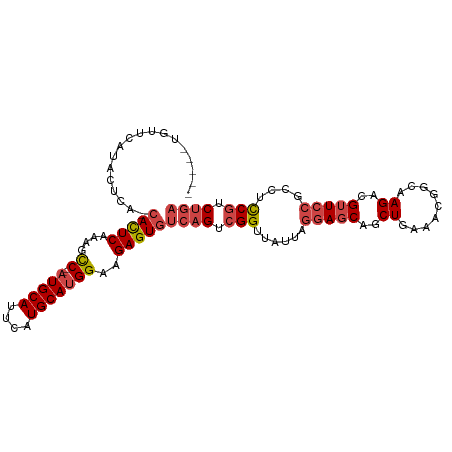
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:55 2011