| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,714,579 – 26,714,678 |
| Length | 99 |
| Max. P | 0.988104 |

| Location | 26,714,579 – 26,714,678 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 66.90 |
| Shannon entropy | 0.62563 |
| G+C content | 0.49890 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.71 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
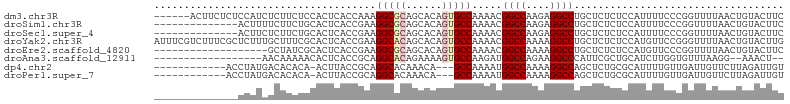
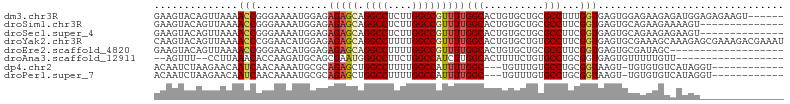
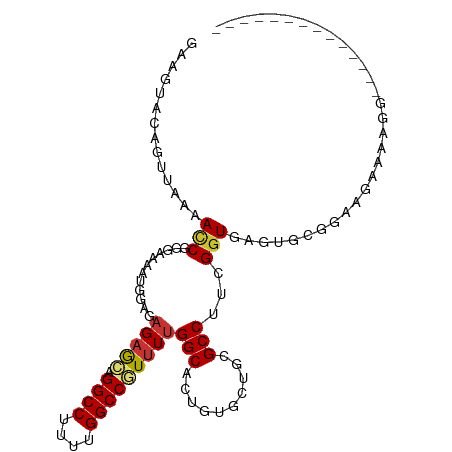
>dm3.chr3R 26714579 99 + 27905053 ------ACUUCUCUCCAUCUCUUCUCCACUCACCAAAGGCGCAGCACAGUGCCAAAACGGCCAAGAGGCCUGCUCUCUCCAUUUUCCCGGUUUUAACUGUACUUC ------.........................(((...(((((......))))).....((((....))))..................))).............. ( -18.80, z-score = -1.11, R) >droSim1.chr3R 26355538 91 + 27517382 --------------ACUUUUCUUCUGCACUCACCGAAGGCGCAGCACAGUGCCAAAACGGCCAAGAGGCCUGCUCUCUCCAUUUUCCCGGUUUUAACUGUACUUC --------------..........((((...((((..(((((......))))).....((((....)))).................))))......)))).... ( -21.10, z-score = -0.77, R) >droSec1.super_4 5540332 91 + 6179234 --------------ACUUCUCUUCUGCACUCACCGAAGGCGCAGCACAGUGCCAAAACGGCCAAGAGGCCUGCUCUCUCCAUUUUCCCGGUUUUAACUGUACUUC --------------..........((((...((((..(((((......))))).....((((....)))).................))))......)))).... ( -21.10, z-score = -0.74, R) >droYak2.chr3R 27638359 105 + 28832112 AUUUCGUCUUUCGCUCUUUGCUUUCGCACUCACCGAAGGCACAGCACAGUGCCAAAACGGCCAAAAGGCCUGCUCUCUCCAUGUUCCGGGUUUUAACUGUACUUG ............(((...((((((((.......)))))))).)))(((((...(((((((((....))))........((.......))))))).)))))..... ( -27.90, z-score = -1.81, R) >droEre2.scaffold_4820 9194518 86 - 10470090 -------------------GCUAUCGCACUCACCGAAGGCGCAGCACAGUGCCAAAACGGCCAAAAGGCCUGCUCUCUCCAUGUUCCCGGUUUUAACUGUACUUC -------------------((....))....((((..(((((......))))).....((((....)))).................)))).............. ( -21.50, z-score = -0.76, R) >droAna3.scaffold_12911 1122170 83 - 5364042 ------------------AACAAAAACACUCACCGCAGGCACAGAAAAGUGCCAAGAUGGCCAGAAGGCCCAUUCGCUGCAUCUUGGUGUUUAAGG--AAACU-- ------------------.....(((((((....((((((((......)))))..((.((((....))))...))..))).....))))))).((.--...))-- ( -26.80, z-score = -2.54, R) >dp4.chr2 30336695 89 - 30794189 ------------ACCUAUGACACACA-ACUUACCGCAGGCACAAACA---GCCAAAAUGGCCAAAAGGCCAGCUCUGCGCAUUUUGUUGAUUGUUCUUAGAUUGU ------------..(((.((((..((-((....(((((((.......---)))....(((((....)))))....))))......))))..))))..)))..... ( -24.10, z-score = -1.75, R) >droPer1.super_7 1205123 89 - 4445127 ------------ACCUAUGACACACA-ACUUACCGCAGGCACAAACA---GCCAAAAUGGCCAAAAGGCCAGCUCUGCGCAUUUUGUUGAUUGUUCUUAGAUUGU ------------..(((.((((..((-((....(((((((.......---)))....(((((....)))))....))))......))))..))))..)))..... ( -24.10, z-score = -1.75, R) >consensus ______________CCAUCACUUACGCACUCACCGAAGGCACAGCACAGUGCCAAAACGGCCAAAAGGCCUGCUCUCUCCAUUUUCCCGGUUUUAACUGUACUUC .....................................(((((......))))).....((((....))))................................... (-14.34 = -14.71 + 0.38)
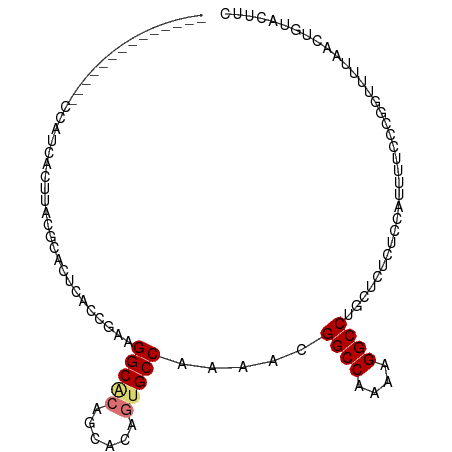
| Location | 26,714,579 – 26,714,678 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 66.90 |
| Shannon entropy | 0.62563 |
| G+C content | 0.49890 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -13.97 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26714579 99 - 27905053 GAAGUACAGUUAAAACCGGGAAAAUGGAGAGAGCAGGCCUCUUGGCCGUUUUGGCACUGUGCUGCGCCUUUGGUGAGUGGAGAAGAGAUGGAGAGAAGU------ ..((((((((.....(((......)))..(((((.((((....)))))))))...)))))))).((((...))))........................------ ( -25.20, z-score = 0.05, R) >droSim1.chr3R 26355538 91 - 27517382 GAAGUACAGUUAAAACCGGGAAAAUGGAGAGAGCAGGCCUCUUGGCCGUUUUGGCACUGUGCUGCGCCUUCGGUGAGUGCAGAAGAAAAGU-------------- ..((((((((.....(((......)))..(((((.((((....)))))))))...))))))))(((((......).))))...........-------------- ( -27.50, z-score = -0.59, R) >droSec1.super_4 5540332 91 - 6179234 GAAGUACAGUUAAAACCGGGAAAAUGGAGAGAGCAGGCCUCUUGGCCGUUUUGGCACUGUGCUGCGCCUUCGGUGAGUGCAGAAGAGAAGU-------------- ..((((((((.....(((......)))..(((((.((((....)))))))))...))))))))(((((......).))))...........-------------- ( -27.50, z-score = -0.44, R) >droYak2.chr3R 27638359 105 - 28832112 CAAGUACAGUUAAAACCCGGAACAUGGAGAGAGCAGGCCUUUUGGCCGUUUUGGCACUGUGCUGUGCCUUCGGUGAGUGCGAAAGCAAAGAGCGAAAGACGAAAU ...(((((((......(((((((.((.......))((((....)))))))))))......))))))).((((.(...(((....)))...).))))......... ( -34.10, z-score = -1.85, R) >droEre2.scaffold_4820 9194518 86 + 10470090 GAAGUACAGUUAAAACCGGGAACAUGGAGAGAGCAGGCCUUUUGGCCGUUUUGGCACUGUGCUGCGCCUUCGGUGAGUGCGAUAGC------------------- ..((((((((.....(((......)))..(((((.((((....)))))))))...))))))))(((((......).))))......------------------- ( -27.50, z-score = -0.52, R) >droAna3.scaffold_12911 1122170 83 + 5364042 --AGUUU--CCUUAAACACCAAGAUGCAGCGAAUGGGCCUUCUGGCCAUCUUGGCACUUUUCUGUGCCUGCGGUGAGUGUUUUUGUU------------------ --.....--.(..((((((((...((((.......((((....)))).....(((((......))))))))).)).))))))..)..------------------ ( -28.20, z-score = -2.16, R) >dp4.chr2 30336695 89 + 30794189 ACAAUCUAAGAACAAUCAACAAAAUGCGCAGAGCUGGCCUUUUGGCCAUUUUGGC---UGUUUGUGCCUGCGGUAAGU-UGUGUGUCAUAGGU------------ ...(((((.((.((....((((..(((((((.(((((((....))))).....((---.....)))))))).)))..)-))).)))).)))))------------ ( -26.50, z-score = -0.87, R) >droPer1.super_7 1205123 89 + 4445127 ACAAUCUAAGAACAAUCAACAAAAUGCGCAGAGCUGGCCUUUUGGCCAUUUUGGC---UGUUUGUGCCUGCGGUAAGU-UGUGUGUCAUAGGU------------ ...(((((.((.((....((((..(((((((.(((((((....))))).....((---.....)))))))).)))..)-))).)))).)))))------------ ( -26.50, z-score = -0.87, R) >consensus GAAGUACAGUUAAAACCGGGAAAAUGGAGAGAGCAGGCCUUUUGGCCGUUUUGGCACUGUGCUGCGCCUUCGGUGAGUGCGGAAGAAAAGG______________ ..............(((............(((((.((((....)))))))))(((.(......).)))...)))............................... (-13.97 = -14.42 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:25 2011