
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,668,508 – 26,668,615 |
| Length | 107 |
| Max. P | 0.831421 |

| Location | 26,668,508 – 26,668,615 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.36170 |
| G+C content | 0.54956 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.82 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811163 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
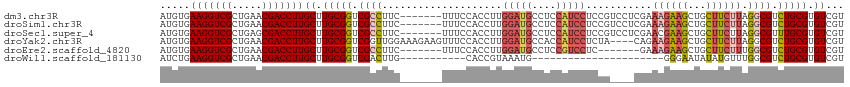
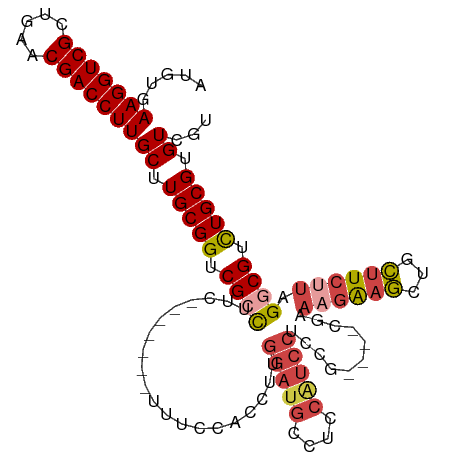
>dm3.chr3R 26668508 107 + 27905053 AUGUGAAGGUCGCUGAACGACCUUGCUUGCGGUCGCCUUC-------UUUCCACCUUGGAUGCCUCCAUCCUCCGUCCUCGAAAGAAGCUGCUUCUUAGGCGUCUGCGUGUCGU .....(((((((.....)))))))((.(((((.(((((((-------((((.((...(((((....)))))...))....))))))...........))))).))))).))... ( -34.40, z-score = -1.86, R) >droSim1.chr3R 26309313 107 + 27517382 AUGUGAAGGUCGCUGAACGACCUUGCUUGCGGUCGCCUUC-------UUUCCACCUUGGAUGCCUCCAUCCUCCGUCCUCGAAAGAAGCUGCUUCUUAGGCGUCUGCGUGUCGU .....(((((((.....)))))))((.(((((.(((((((-------((((.((...(((((....)))))...))....))))))...........))))).))))).))... ( -34.40, z-score = -1.86, R) >droSec1.super_4 5493711 107 + 6179234 AUGUGAAGGUCGCUGAGCGACCUUGCUUGCGGUCGCCUUC-------UUUCCACCUUGGAUGCCUCCAUCCUCCGUCCUCGAACGAAGCUGCUUCUUAGGCGUUUGCGUGUCGU .....((((((((...))))))))((.(((((.(((((..-------....((.((((((((....)))))..(((......)))))).))......))))).))))).))... ( -32.90, z-score = -0.85, R) >droYak2.chr3R 27590777 110 + 28832112 AUGUGAAGGUCGCUGAACGACCUUGCUUGCGGUCGGUUGGAAAGAAGUUUCCACCUUGGAUGCCACCAUCCUCUA----CAGAAGAAGCUGCUUCUUAGGCGUCUGCGUGUCGU .....(((((((.....)))))))((.(((((.((((.(((((....))))))))..(((((....)))))((((----.(((((......))))))))).).))))).))... ( -34.70, z-score = -0.56, R) >droEre2.scaffold_4820 9142700 100 - 10470090 AUGUGAAGGUCGCUGAACGACCUUGCUUGCGGUCGCCUUC-------UUUCCACCUUGGAUGCCUCCGUCCUC-------GAAAGAAGCUGCUUCUUUGGCGUCUGCGUGUCGU .....(((((((.....)))))))((.(((((.((((...-------..........(((((....)))))..-------.(((((((...))))))))))).))))).))... ( -34.30, z-score = -2.28, R) >droWil1.scaffold_181130 10244426 81 + 16660200 AUCUGAAGGUCGCUGAACGACCUUGCUUGCGGUCGACUUG-----------CACCGUAAAUG----------------------GGGAAUAUAUGUUUGGCGUCUGCGUGUCGU ....((.((.(((..((((.((((..(((((((.......-----------.)))))))..)----------------------)))......))))..))).)).....)).. ( -23.70, z-score = -0.43, R) >consensus AUGUGAAGGUCGCUGAACGACCUUGCUUGCGGUCGCCUUC_______UUUCCACCUUGGAUGCCUCCAUCCUCCG____CGAAAGAAGCUGCUUCUUAGGCGUCUGCGUGUCGU .....(((((((.....)))))))((.(((((.((((....................(((((....)))))...........((((((...)))))).)))).))))).))... (-25.62 = -26.82 + 1.20)

| Location | 26,668,508 – 26,668,615 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.36170 |
| G+C content | 0.54956 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -24.47 |
| Energy contribution | -26.30 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831421 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
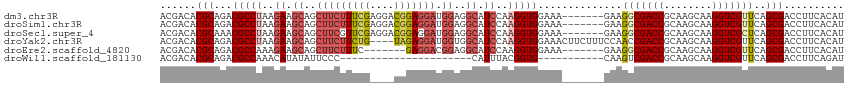
>dm3.chr3R 26668508 107 - 27905053 ACGACACGCAGACGCCUAAGAAGCAGCUUCUUUCGAGGACGGAGGAUGGAGGCAUCCAAGGUGGAAA-------GAAGGCGACCGCAAGCAAGGUCGUUCAGCGACCUUCACAU .......((.(.(((((.....((..(((((((((....))))))).))..)).(((.....)))..-------..))))).).))..(.(((((((.....)))))))).... ( -34.90, z-score = -1.49, R) >droSim1.chr3R 26309313 107 - 27517382 ACGACACGCAGACGCCUAAGAAGCAGCUUCUUUCGAGGACGGAGGAUGGAGGCAUCCAAGGUGGAAA-------GAAGGCGACCGCAAGCAAGGUCGUUCAGCGACCUUCACAU .......((.(.(((((.....((..(((((((((....))))))).))..)).(((.....)))..-------..))))).).))..(.(((((((.....)))))))).... ( -34.90, z-score = -1.49, R) >droSec1.super_4 5493711 107 - 6179234 ACGACACGCAAACGCCUAAGAAGCAGCUUCGUUCGAGGACGGAGGAUGGAGGCAUCCAAGGUGGAAA-------GAAGGCGACCGCAAGCAAGGUCGCUCAGCGACCUUCACAU ......(((...(((((.........(((((((....)))))))..((((....)))))))))....-------...(((((((........)))))))..))).......... ( -36.30, z-score = -1.76, R) >droYak2.chr3R 27590777 110 - 28832112 ACGACACGCAGACGCCUAAGAAGCAGCUUCUUCUG----UAGAGGAUGGUGGCAUCCAAGGUGGAAACUUCUUUCCAACCGACCGCAAGCAAGGUCGUUCAGCGACCUUCACAU .......((....((((..((.((.((((((((..----..))))).))).)).))..))))(((((....)))))........))..(.(((((((.....)))))))).... ( -32.10, z-score = -0.83, R) >droEre2.scaffold_4820 9142700 100 + 10470090 ACGACACGCAGACGCCAAAGAAGCAGCUUCUUUC-------GAGGACGGAGGCAUCCAAGGUGGAAA-------GAAGGCGACCGCAAGCAAGGUCGUUCAGCGACCUUCACAU .......((.(.((((.........((((((.((-------...)).)))))).(((.....)))..-------...)))).).))..(.(((((((.....)))))))).... ( -31.00, z-score = -1.41, R) >droWil1.scaffold_181130 10244426 81 - 16660200 ACGACACGCAGACGCCAAACAUAUAUUCCC----------------------CAUUUACGGUG-----------CAAGUCGACCGCAAGCAAGGUCGUUCAGCGACCUUCAGAU .((((..(((..((................----------------------......)).))-----------)..)))).......(.(((((((.....)))))))).... ( -17.45, z-score = -1.10, R) >consensus ACGACACGCAGACGCCUAAGAAGCAGCUUCUUUCG____CGGAGGAUGGAGGCAUCCAAGGUGGAAA_______GAAGGCGACCGCAAGCAAGGUCGUUCAGCGACCUUCACAU ......(((...(((((..((.((..(((((((((....))))))).))..)).))..)))))..............(((((((........)))))))..))).......... (-24.47 = -26.30 + 1.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:22 2011