
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,666,400 – 26,666,468 |
| Length | 68 |
| Max. P | 0.848458 |

| Location | 26,666,400 – 26,666,468 |
|---|---|
| Length | 68 |
| Sequences | 13 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 90.59 |
| Shannon entropy | 0.21531 |
| G+C content | 0.50226 |
| Mean single sequence MFE | -16.42 |
| Consensus MFE | -12.71 |
| Energy contribution | -12.87 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
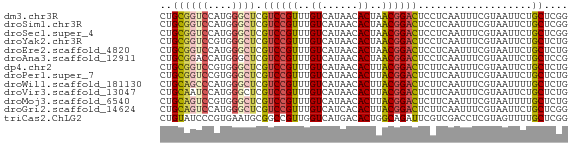
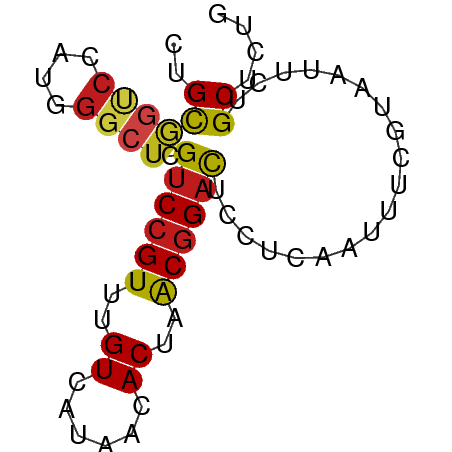
>dm3.chr3R 26666400 68 - 27905053 CUGCGGUCCAUGGGCUCGUCCGUUUGUCAUAACACUAACGGACUCCUCAAUUUCGUAAUUCUGCUCGG .(((((....((((...(((((((.((......)).)))))))..))))...)))))........... ( -17.00, z-score = -2.00, R) >droSim1.chr3R 26307337 68 - 27517382 CUGCGGUCCAUGGGCUCGUCCGUUUGUCAUAACACUAACGGACUCCUCAAUUUCGUAAUUCUGCUCGG .(((((....((((...(((((((.((......)).)))))))..))))...)))))........... ( -17.00, z-score = -2.00, R) >droSec1.super_4 5491699 68 - 6179234 CUGCGGUCCAUGGGCUCGUCCGUUUGUCAUAACACUAACGGACUCCUCAAUUUCGUAAUUCUGCUCGG .(((((....((((...(((((((.((......)).)))))))..))))...)))))........... ( -17.00, z-score = -2.00, R) >droYak2.chr3R 27588596 68 - 28832112 CUGCGGUCCGUGGGCUCGUCCGUUUGUCAUAACACUAACGGACUCCUCAAUUUCGUAAUUCUGCUCUG .(((((....((((...(((((((.((......)).)))))))..))))...)))))........... ( -18.10, z-score = -2.76, R) >droEre2.scaffold_4820 9140570 68 + 10470090 CUGCGGUCCAUGGGCUCGUCCGUUUGUCAUAACACUAACGGACUCCUCAAUUUCGUAAUUCUGCUCUG .(((((....((((...(((((((.((......)).)))))))..))))...)))))........... ( -17.00, z-score = -2.71, R) >droAna3.scaffold_12911 1052899 68 + 5364042 CUGCGGACCAUGGGCUCGUCCGUUUGUCAUAACACUAACGGACUCCUCAAUUUCGUAAUUCUGCUCCG .((((((...((((...(((((((.((......)).)))))))..))))..))))))........... ( -18.00, z-score = -2.53, R) >dp4.chr2 30280257 68 + 30794189 CUGCGGUCCGUGGGCUCGUCCGUUUGUCAUAACACUUACGGACUCUUCAAUUUCGUAAUUCUGCUCUG .(((((((....)))..((((((..((......))..))))))..........))))........... ( -16.20, z-score = -2.12, R) >droPer1.super_7 1148187 68 + 4445127 CUGCGGUCCGUGGGCUCGUCCGUUUGUCAUAACACUUACGGACUCUUCAAUUUCGUAAUUCUGCUCUG .(((((((....)))..((((((..((......))..))))))..........))))........... ( -16.20, z-score = -2.12, R) >droWil1.scaffold_181130 10241873 68 - 16660200 CUGCAGCCCAUGGGCUCGUCCGUUUGUCAUAACACUUACGGACUCUUCAAUUUCGUAAUUUUGCUCUG ..((((((....)))..((((((..((......))..))))))..................))).... ( -18.60, z-score = -3.60, R) >droVir3.scaffold_13047 11493819 68 - 19223366 CUGCAAUCCAUGGGCUCGUCCGUUUGUCAUAACACUUACGGACUCUUCAAUUUCGUAAUUCUGCUCUG ...........((((..((((((..((......))..)))))).....((((....))))..)))).. ( -13.40, z-score = -2.33, R) >droMoj3.scaffold_6540 3889864 68 + 34148556 CUGCAGUCCGUGGGCUCGUCCGUUUGUCAUAACACUUACGGACUCUUCAAUUUCGUAAUUUUGCUCUG ..((((((....)))..((((((..((......))..))))))..................))).... ( -15.90, z-score = -2.29, R) >droGri2.scaffold_14624 1542509 68 - 4233967 CUGCAGUCCAUGGGCUCGUCCGUUUGUCAUCACACUUACGGACUCUUCAAUUUCGUAAUUCUGCUCGG ..((((((....)))..((((((..((......))..))))))..................))).... ( -15.80, z-score = -2.16, R) >triCas2.ChLG2 6375754 68 + 12900155 CUGUAUCCCGUGAAUGCGGCCGUUGGUCAUGACACUGGCAGAUUCGUCGACCUCGUAGUUUUGCUCGG ........(((((..((....))...)))))...((((((((.....((....))....))))).))) ( -13.30, z-score = 1.85, R) >consensus CUGCGGUCCAUGGGCUCGUCCGUUUGUCAUAACACUAACGGACUCCUCAAUUUCGUAAUUCUGCUCUG ..((((((....)))).((((((..((......))..))))))...................)).... (-12.71 = -12.87 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:21 2011