
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,663,100 – 26,663,205 |
| Length | 105 |
| Max. P | 0.555927 |

| Location | 26,663,100 – 26,663,205 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.79 |
| Shannon entropy | 0.57904 |
| G+C content | 0.42041 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -12.30 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
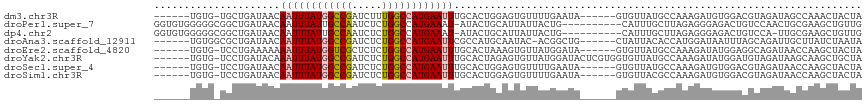
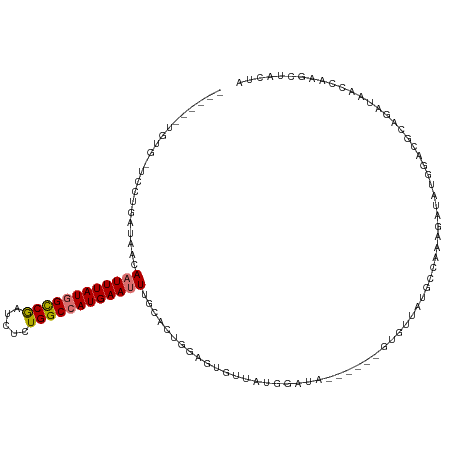
>dm3.chr3R 26663100 105 + 27905053 ------UGUG-UGCUGAUAACAAUUUAUGGCCGAUCUUUGGCCAUGAAUUUGCACUGGAGUGUUUUGAAUA------GUGUUAUGCCAAAGAUGUGGACGUAGAUAGCCAAACUACUA ------...(-(((.......(((((((((((((...))))))))))))).))))...((((.((((..((------...((((((((......))).))))).))..)))).)))). ( -30.00, z-score = -2.39, R) >droPer1.super_7 1144504 107 - 4445127 GGUGUGGGGGCGGCUGAUAACAAUUUAUUGCCAAUCUCUGGCCAUGAAAU-AUACUGCAUUAUUACUG----------CAUUUGCUUAGAGGGAGACUGUCCAACUGCGAAGCUGUUG ........(((((((........(((((.((((.....)))).)))))..-....((((.......))----------)).((((..((..(((.....)))..))))))))))))). ( -23.50, z-score = 1.28, R) >dp4.chr2 30276559 106 - 30794189 GGUGUGGGGGCGGCUGAUAACAAUUUAUUGCCAAUCUCUGGCCAUGAAAU-AUACUGCAUUAUUACUG----------CAUUUGCUUAGAGGGAGACUGUCCA-UUGCGAAGCUGUUG ........(((((((........(((((.((((.....)))).)))))..-....((((.......))----------)).((((.....((....)).....-..))))))))))). ( -23.70, z-score = 1.24, R) >droAna3.scaffold_12911 1049440 105 - 5364042 ------UGUGGCGCUGAUAACAAUUUAUGGCCGAUCUCUGGCCAUGAAUUCGCCAUGCAAUAC-ACGGCUG------CUAUUACACCAUGGAUAAUUUAGCAGAUUGCUUAUCUAAUA ------.((((((........((((((((((((.....))))))))))))))))))(((((..-....(((------(((.....(....)......))))))))))).......... ( -31.50, z-score = -2.54, R) >droEre2.scaffold_4820 9137186 105 - 10470090 ------UGUG-UCCUGAAAAAAAUUUAUGGUCGCUCUCUGGCCAUGAAUUUGCACUAAAGUGUUAUGGAUA------GUGUUAUGCCAAAGAUAUGGAGGCAGAUAACCAAGCUACUA ------..((-(((......(((((((((((((.....)))))))))))))((((....))))...)))))------(.((((((((...........))))..)))))......... ( -26.70, z-score = -1.41, R) >droYak2.chr3R 27585193 111 + 28832112 ------UGUG-UCCUGAUACAAAUUUAUGGCCGAUCUCUGGCCAUGAAUUUGCACUAGAGUGUUAUGGAUACUCGUGGUGUUAUGCCAAAGAUAUGGAUGUAGAUAAGCAAGCUGCUA ------((..-(((.....((((((((((((((.....))))))))))))))((((((((((((...))))))).)))))...............)))..))....(((.....))). ( -37.20, z-score = -3.31, R) >droSec1.super_4 5488345 105 + 6179234 ------UGUG-UCCUGAUAACAAUUUAUGGCCGAUCUCUGGCCAUGAAUUUGCACUGGAGUGUUUUGAAUA------GUGUUAUGCCAAAGAUGUGGACGUAGAUAACCAAGCUACUA ------...(-(((.......((((((((((((.....)))))))))))).((((((............))------))))..............))))((((.........)))).. ( -26.70, z-score = -1.32, R) >droSim1.chr3R 26303986 105 + 27517382 ------UGUG-UCCUGAUAACAAUUUAUGGCCGAUCUCUGGCCAUGAAUUUGCACUGGAGUGUUUUGAAUA------GUGUUACGCCAAAGAUGUGGACGUAGAUAACCAAGCUACUA ------....-(((.......((((((((((((.....))))))))))))......)))(((.((((..((------...((((((((......))).))))).))..)))).))).. ( -28.12, z-score = -1.70, R) >consensus ______UGUG_UCCUGAUAACAAUUUAUGGCCGAUCUCUGGCCAUGAAUUUGCACUGGAGUGUUAUGGAUA______GUGUUAUGCCAAAGAUAUGGACGCAGAUAACCAAGCUACUA .....................((((((((((((.....)))))))))))).................................................................... (-12.30 = -12.50 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:20 2011