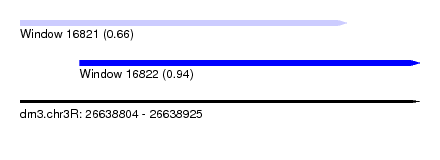
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,638,804 – 26,638,925 |
| Length | 121 |
| Max. P | 0.941941 |

| Location | 26,638,804 – 26,638,903 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 68.04 |
| Shannon entropy | 0.59428 |
| G+C content | 0.49710 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -10.84 |
| Energy contribution | -11.84 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26638804 99 + 27905053 UGCCUCUGCUUUUCACACGCAAUGUGCGGCC-UUGAACUUGAGCAAUUGCUU-UCAUUUCAGUGUUUCAGGGCUCGCAAAGCUCAUAA-GUCAAGGUCACCU--- .((((..((((.......((....(((((((-(((((...((((....))))-.(((....))).)))))))).))))..))....))-))..)))).....--- ( -27.70, z-score = -0.95, R) >droSim1.chr3R 26280242 79 + 27517382 UGCCUCUGCUUUUCACACGCAAUGUGCGGCC-UUGAACUUGAGCAAUUGCUU-UC--------------------GCAAAGCUCAUAA-GUCAAGGUCACCU--- .(((..(((.........)))..).))((((-((((...(((((..((((..-..--------------------)))).)))))...-.))))))))....--- ( -27.60, z-score = -3.31, R) >droSec1.super_4 5464925 79 + 6179234 UGCCUCUGCUUUUCACACGCAAUGUGCGGCC-UUGAACUUGAGCAAUUGCUU-UC--------------------GCAAAGCUCAUAA-GUCAAGGUCACCU--- .(((..(((.........)))..).))((((-((((...(((((..((((..-..--------------------)))).)))))...-.))))))))....--- ( -27.60, z-score = -3.31, R) >droYak2.chr3R 27561463 99 + 28832112 UGCCUCCGUAUUUCACACGCAAUGUGCGGCC-UUGAACUUGAGCAAUUGCUU-UCACUUCGGUGUUUCAGGGCUAGCAAAGCUCAUAA-GUCAAGGUCACCU--- ..................((.....))((((-((((...(((((..(((((.-.(((....)))..........))))).)))))...-.))))))))....--- ( -29.90, z-score = -1.75, R) >droEre2.scaffold_4820 9112701 99 - 10470090 UGCCUUAGUUUUUCACACGCAAUGUGCGGCC-UUGAACUUGGGCAAUUGCGU-UCACUUCAGCGUUUCAGGGCUAGCAAAGCUCAUAA-GUCAAGGUCACCU--- ..................((.....))((((-((((...(((((..(((((.-...)...(((........))).)))).)))))...-.))))))))....--- ( -25.50, z-score = -0.04, R) >dp4.chr2 30246023 102 - 30794189 --UGUUCUCCUCUCACACGCCUUUUGUGGCC-UUGAACUUGGACAACUUCGAGUUGCCGUUACUCGAGAGGCUUUGUAACGCACCUCCCGGCAAGGUCAAUGCGC --...............(((......(((((-((.((((((((....))))))))((((......(((..((........))..))).)))))))))))..))). ( -27.80, z-score = -0.19, R) >droPer1.super_7 1113215 103 - 4445127 --UGUUCUCCUCUCACACGCCUUUUGUGGCCCUUGAACUUGGACAACUUCGAGUUGCCGUUACUCGAGAGGCUUUGUAACGCACCUCCCGUCAAGGUCAGUGCGU --......((((((....(((......)))....((...(((.((((.....)))))))....)))))))).......((((((...((.....))...)))))) ( -30.90, z-score = -1.19, R) >consensus UGCCUCCGCUUUUCACACGCAAUGUGCGGCC_UUGAACUUGAGCAAUUGCUU_UCACUUC_GCGUUUCAGGGCU_GCAAAGCUCAUAA_GUCAAGGUCACCU___ ..................((.....))((((.((((...(((((...(((.........................)))..))))).....))))))))....... (-10.84 = -11.84 + 1.00)


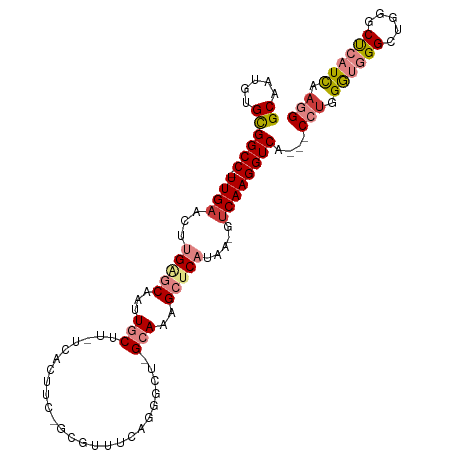
| Location | 26,638,822 – 26,638,925 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 70.64 |
| Shannon entropy | 0.54615 |
| G+C content | 0.54391 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -17.23 |
| Energy contribution | -18.58 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.941941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26638822 103 + 27905053 GCAAUGUGCGGCC-UUGAACUUGAGCAAUUGCUU-UCAUUUCAGUGUUUCAGGGCUCGCAAAGCUCAUAA-GUCAAGGUCA---CCUGGGUGGGCUGGGCUCAUCAAGG ((.....))((((-((((.(((((((....))).-................(((((.....))))).)))-))))))))).---(((.(((((((...))))))).))) ( -34.90, z-score = -1.36, R) >droSim1.chr3R 26280260 83 + 27517382 GCAAUGUGCGGCC-UUGAACUUGAGCAAUUGCUU-UC--------------------GCAAAGCUCAUAA-GUCAAGGUCA---CCUGGGUGGGCUGGGCCCAUCAAGG ((.....))((((-((((...(((((..((((..-..--------------------)))).)))))...-.)))))))).---(((.(((((((...))))))).))) ( -38.90, z-score = -4.12, R) >droSec1.super_4 5464943 83 + 6179234 GCAAUGUGCGGCC-UUGAACUUGAGCAAUUGCUU-UC--------------------GCAAAGCUCAUAA-GUCAAGGUCA---CCUGGGUGGGCUGGGCUCAUCAAGG ((.....))((((-((((...(((((..((((..-..--------------------)))).)))))...-.)))))))).---(((.(((((((...))))))).))) ( -35.90, z-score = -3.77, R) >droYak2.chr3R 27561481 103 + 28832112 GCAAUGUGCGGCC-UUGAACUUGAGCAAUUGCUU-UCACUUCGGUGUUUCAGGGCUAGCAAAGCUCAUAA-GUCAAGGUCA---CCUGGGUGGGCUGGGCUCAUCAAGG ((.....))((((-((((...(((((..(((((.-.(((....)))..........))))).)))))...-.)))))))).---(((.(((((((...))))))).))) ( -40.70, z-score = -2.86, R) >droEre2.scaffold_4820 9112719 103 - 10470090 GCAAUGUGCGGCC-UUGAACUUGGGCAAUUGCGU-UCACUUCAGCGUUUCAGGGCUAGCAAAGCUCAUAA-GUCAAGGUCA---CCUGGGUGGGCCGGGCUCAUCAAGG ((.....))((((-((((...(((((..(((((.-...)...(((........))).)))).)))))...-.)))))))).---(((.(((((((...))))))).))) ( -36.30, z-score = -1.33, R) >dp4.chr2 30246039 108 - 30794189 GCCUUUUGUGGCC-UUGAACUUGGACAACUUCGAGUUGCCGUUACUCGAGAGGCUUUGUAACGCACCUCCCGGCAAGGUCAAUGCGCGGAGAGGGGGUGCCCAUUCAGG ((((((((((((.-...((((((((....)))))))))))).....))))))))........(((((((((((((.......))).))....))))))))......... ( -36.60, z-score = -0.05, R) >droPer1.super_7 1113231 109 - 4445127 GCCUUUUGUGGCCCUUGAACUUGGACAACUUCGAGUUGCCGUUACUCGAGAGGCUUUGUAACGCACCUCCCGUCAAGGUCAGUGCGUGGAGAGGGGGUGCCCAUUCAGG .(((...(((((((((((...(((.((((.....)))))))....))))).)))........(((((((((..((..((....)).))..).)))))))).)))..))) ( -37.10, z-score = -0.18, R) >consensus GCAAUGUGCGGCC_UUGAACUUGAGCAAUUGCUU_UCACUUC_GCGUUUCAGGGCU_GCAAAGCUCAUAA_GUCAAGGUCA___CCUGGGUGGGCUGGGCUCAUCAAGG ((.....))((((.((((...(((((...(((.........................)))..))))).....))))))))....(((.((((((.....)))))).))) (-17.23 = -18.58 + 1.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:16 2011