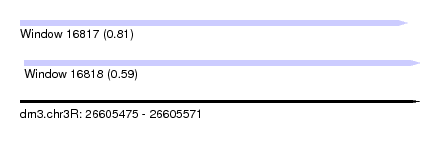
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,605,475 – 26,605,571 |
| Length | 96 |
| Max. P | 0.812361 |

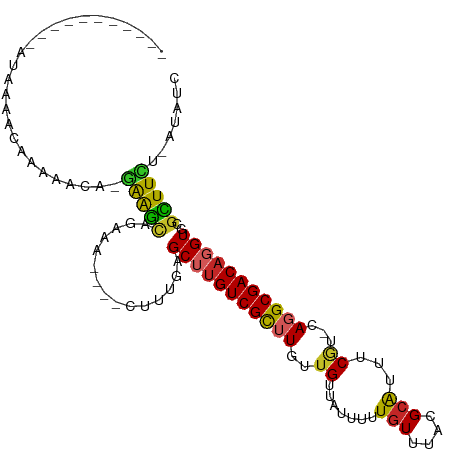
| Location | 26,605,475 – 26,605,568 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.42 |
| Shannon entropy | 0.44335 |
| G+C content | 0.39113 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -13.55 |
| Energy contribution | -13.06 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26605475 93 + 27905053 -----------AUAAAACAAAAACA-UAAGCAGCAA----CUUUGAGCUUGUCGCUUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUCU-AUAUC -----------...(((((((((((-.((((.((((----........)))).))))..)))...)))))))).(((((((.((-(....)))))))))).....-..... ( -22.20, z-score = -1.63, R) >droSim1.chr3R 26245824 93 + 27517382 -----------GUAAAACAAAAACA-GAAGCAGAAA----CUUUGAGCUUGUCGCUUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUCU-AUAUC -----------.............(-(((((...((----(...((((.....))))...)))............((((((.((-(....)))))))))))))))-..... ( -22.50, z-score = -1.15, R) >droSec1.super_4 5423542 92 + 6179234 ------------UAAAACAAAAACA-GAAGCAGAAA----CUUUGAGCUUGUCGCUUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUCU-AUAUC ------------............(-(((((...((----(...((((.....))))...)))............((((((.((-(....)))))))))))))))-..... ( -22.50, z-score = -1.36, R) >droYak2.chr3R 27526934 93 + 28832112 -----------AUUAAACAAAAACA-GAAGCAGCAA----CUUUGAGCUUGUCGCAUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUCU-AUAUC -----------..((((((((((((-(((((.((((----........)))).)).))))))...)))))))))(((((((.((-(....)))))))))).....-..... ( -24.00, z-score = -1.97, R) >droEre2.scaffold_4820 9076665 93 - 10470090 -----------AUUAAACAAAAACA-GAAGCAGAAA----CUUUGAGCUUGUCGCAUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUUU-AUAUC -----------..((((((((((((-((((((.((.----(.....).)).).)).))))))...)))))))))(((((((.((-(....)))))))))).....-..... ( -19.60, z-score = -0.65, R) >droAna3.scaffold_12911 985391 94 - 5364042 -----------AUAAAACAAAAACA-GAAGCAGAAAA---CUUUGAGCUUGUCGCUUGUUGUUAUUUUUGUUUACGCGUUUCGU-CAGGCGACAGGUGCGCUUCU-UUAUC -----------.............(-(((((((....---))..(.((((((((((((.((..((...((....)).))..)).-)))))))))))).)))))))-..... ( -23.80, z-score = -0.92, R) >dp4.chr2 30203726 109 - 30794189 AAGAAUAAAACAGAAAACACAAGCAAGAGGCAGAAGAAAACUUUGAGCUUGUCGCUUGUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGUUUCU-AUAUC .((((..(((((((((((((((((.(.((((.((((....))))..)))).).))))).)))..)))))))))((((((((.((-(....)))))))))))))))-..... ( -33.90, z-score = -3.42, R) >droPer1.super_7 1070542 106 - 4445127 ---AAGAAAACAGAAAACACAAGCAAGAGGCAGAAGAAAACUUUGAGCUUGUCGCUUGUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGUUUCU-AUAUC ---.((((((((((((((((((((.(.((((.((((....))))..)))).).))))).)))..)))))))))((((((((.((-(....))))))))))).)))-..... ( -33.20, z-score = -3.08, R) >droWil1.scaffold_181130 10145674 91 + 16660200 ---------------GGAGAGUGGAUGGGGUGGAACG----GCAAAGCAUGUCGUUUGUUGUUAUUUCUGUUUACGCAUUUCAU-AAGGCGACAGGUGCGUUUUUUAUAUG ---------------.....((((((((((...((((----(((((........)))))))))..))))))))))((((((...-..(....)))))))............ ( -20.00, z-score = -0.27, R) >droVir3.scaffold_13047 11412577 80 + 19223366 -------------------------AAAAAAAACAA-----CUCGAGCAUGUCGUUUGUUGUUAUUUUU-UUUACGGGUUGCCCACAGGCGACCGGUGGGUUUUUUAUAUG -------------------------..(((((((((-----(.(((((.....)))))..)))......-.((((.(((((((....))))))).)))))))))))..... ( -24.30, z-score = -2.72, R) >consensus ___________AUAAAACAAAAACA_GAAGCAGAAA____CUUUGAGCUUGUCGCUUGUUGUUAUUUUUGUUUACGCAUUUCGU_CAGGCGACAGGUGCGCUUCU_AUAUC ..........................(((((...............(((((((((((...........(((....)))........)))))))))))..)))))....... (-13.55 = -13.06 + -0.49)


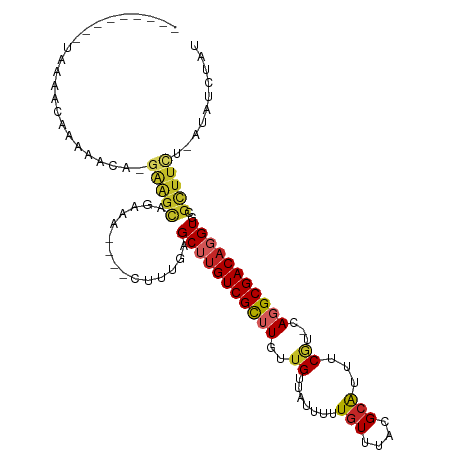
| Location | 26,605,476 – 26,605,571 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Shannon entropy | 0.42586 |
| G+C content | 0.38001 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -12.51 |
| Energy contribution | -12.75 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.588605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26605476 95 + 27905053 ---------UAAAACAAAAACA-UAAGCAGCAA----CUUUGAGCUUGUCGCUUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUCU-AUAUCUAU ---------..(((((((((((-.((((.((((----........)))).))))..)))...)))))))).(((((((.((-(....)))))))))).....-........ ( -22.20, z-score = -1.52, R) >droSim1.chr3R 26245825 95 + 27517382 ---------UAAAACAAAAACA-GAAGCAGAAA----CUUUGAGCUUGUCGCUUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUCU-AUAUCUAU ---------............(-(((((...((----(...((((.....))))...)))............((((((.((-(....)))))))))))))))-........ ( -22.50, z-score = -1.30, R) >droSec1.super_4 5423542 95 + 6179234 ---------UAAAACAAAAACA-GAAGCAGAAA----CUUUGAGCUUGUCGCUUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUCU-AUAUCUAU ---------............(-(((((...((----(...((((.....))))...)))............((((((.((-(....)))))))))))))))-........ ( -22.50, z-score = -1.30, R) >droYak2.chr3R 27526935 95 + 28832112 ---------UUAAACAAAAACA-GAAGCAGCAA----CUUUGAGCUUGUCGCAUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUCU-AUAUCUAU ---------.((((((((((((-(((((.((((----........)))).)).))))))...)))))))))(((((((.((-(....)))))))))).....-........ ( -24.00, z-score = -1.84, R) >droEre2.scaffold_4820 9076666 95 - 10470090 ---------UUAAACAAAAACA-GAAGCAGAAA----CUUUGAGCUUGUCGCAUUUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGCUUUU-AUAUCUAU ---------.((((((((((((-((((((.((.----(.....).)).).)).))))))...)))))))))(((((((.((-(....)))))))))).....-........ ( -19.60, z-score = -0.53, R) >droAna3.scaffold_12911 985392 96 - 5364042 ---------UAAAACAAAAACA-GAAGCAGAAAA---CUUUGAGCUUGUCGCUUGUUGUUAUUUUUGUUUACGCGUUUCGU-CAGGCGACAGGUGCGCUUCU-UUAUCUAU ---------............(-(((((((....---))..(.((((((((((((.((..((...((....)).))..)).-)))))))))))).)))))))-........ ( -23.80, z-score = -0.87, R) >dp4.chr2 30203729 109 - 30794189 AAUAAAACAGAAAACACAAGCAAGAGGCAGAAGAAAACUUUGAGCUUGUCGCUUGUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGUUUCU-AUAUCUAU ....(((((((((((((((((.(.((((.((((....))))..)))).).))))).)))..)))))))))((((((((.((-(....)))))))))))....-........ ( -32.30, z-score = -3.04, R) >droPer1.super_7 1070545 106 - 4445127 ---AAAACAGAAAACACAAGCAAGAGGCAGAAGAAAACUUUGAGCUUGUCGCUUGUUGUUAUUUUUGUUUACGCAUUUCGU-CAGGCGACAGGUGCGUUUCU-AUAUCUAU ---.(((((((((((((((((.(.((((.((((....))))..)))).).))))).)))..)))))))))((((((((.((-(....)))))))))))....-........ ( -32.30, z-score = -2.97, R) >droWil1.scaffold_181130 10145675 93 + 16660200 -------------GAGAGUGGAUGGGGUGGAAC----GGCAAAGCAUGUCGUUUGUUGUUAUUUCUGUUUACGCAUUUCAU-AAGGCGACAGGUGCGUUUUUUAUAUGUAU -------------....((((((((((...(((----((((((........)))))))))..))))))))))((((.....-((((((.(....)))))))....)))).. ( -22.20, z-score = -0.91, R) >droVir3.scaffold_13047 11412578 82 + 19223366 ----------------------------AAAAAAACAACUCGAGCAUGUCGUUUGUUGUUAUUUUU-UUUACGGGUUGCCCACAGGCGACCGGUGGGUUUUUUAUAUGUAU ----------------------------.((((((((((.(((((.....)))))..)))......-.((((.(((((((....))))))).)))))))))))........ ( -24.30, z-score = -2.57, R) >consensus _________UAAAACAAAAACA_GAAGCAGAAA____CUUUGAGCUUGUCGCUUGUUGUUAUUUUUGUUUACGCAUUUCGU_CAGGCGACAGGUGCGCUUCU_AUAUCUAU .......................(((((...............(((((((((((...........(((....)))........)))))))))))..))))).......... (-12.51 = -12.75 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:13 2011