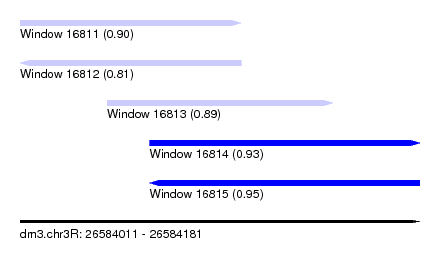
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,584,011 – 26,584,181 |
| Length | 170 |
| Max. P | 0.952622 |

| Location | 26,584,011 – 26,584,105 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Shannon entropy | 0.45068 |
| G+C content | 0.43701 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -11.44 |
| Energy contribution | -11.86 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26584011 94 + 27905053 GAAGCAAGAAUAGAGUGCGG---GAUGUUCGAGGGGAGGGGGGGAAACGCAU-CAAAACCUCACUCGAGAUACAAUUGUAUCU-UUGAAAUUCAAACUG .............(((...(---(((.((((((..((((.(.(....).)..-.....)))).)))(((((((....))))))-).)))))))..))). ( -24.30, z-score = -1.87, R) >droSim1.chr3R 26223438 90 + 27517382 GAAGCAAGAAUAGAGUGCGG---GAUGUUCGAGGGGUGGGG----AACGCAU-CAAAACCUCACCCGAGAUACAAUUGUAUCU-UUGAAUUUCAAACUG .............(((....---((.((((((.((((((((----.......-.....)))))))).((((((....))))))-)))))).))..))). ( -26.60, z-score = -2.29, R) >droSec1.super_4 5402564 90 + 6179234 GAAGCAAGAAUAGAGUGCGG---GAUGUUCGAGGGGUGGGG----AACGCAU-CAAAACCUCACCCGAGAUACAAUUGUAUCU-UUGAAUUUCAAACUG .............(((....---((.((((((.((((((((----.......-.....)))))))).((((((....))))))-)))))).))..))). ( -26.60, z-score = -2.29, R) >droYak2.chr3R 27504930 92 + 28832112 GAAGCAAGAAUAGAGUGCGUUGCGAUGUUCGAGGGGGGAA-----AACGCAG-CAAAACCUCACUCUAGAUACAAUUGUAUCU-UUGAAUUUCCAACUG ...(((.........)))((((.((.(((((((((((...-----.......-.....)))).))).((((((....))))))-..)))).)))))).. ( -22.96, z-score = -1.26, R) >droEre2.scaffold_4820 9055061 88 - 10470090 AAAACAGAAAUAGAGUGCGG---GAUGUUUGAGGGGGGA------AACGCAU-CAAAACCUCACUCGAGAUACAAUUGUAUCU-UUGAAUUUCAAGCUG ......(((((.(((((.((---....(((((..(.(..------..).).)-)))).)).)))))(((((((....))))))-)...)))))...... ( -28.40, z-score = -4.26, R) >droAna3.scaffold_12911 963784 83 - 5364042 ----------------GAAGAGCGAUUUAAGGGAGGGGGUAGUAGAACGCAUUCGAAACCUUAUUUGAGAUACGAUUGUAUCUAUUAGGUUGGAAACAG ----------------(((..(((.((((..(.......)..)))).))).)))..(((((......((((((....))))))...)))))(....).. ( -16.60, z-score = -1.23, R) >consensus GAAGCAAGAAUAGAGUGCGG___GAUGUUCGAGGGGGGGGG____AACGCAU_CAAAACCUCACUCGAGAUACAAUUGUAUCU_UUGAAUUUCAAACUG .......................((.(((((((((((((...................)))).))).((((((....)))))).)))))).))...... (-11.44 = -11.86 + 0.42)
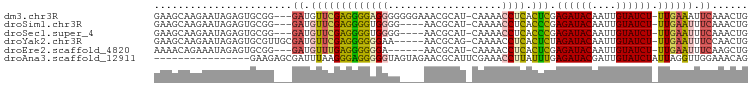
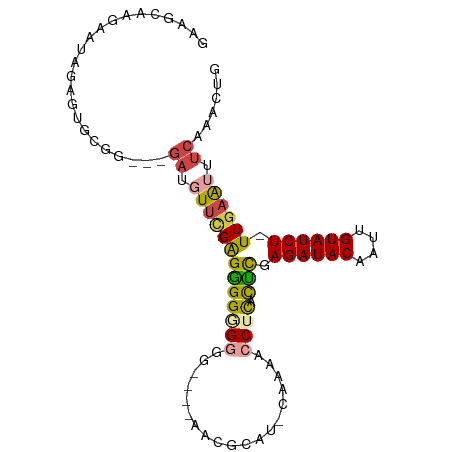
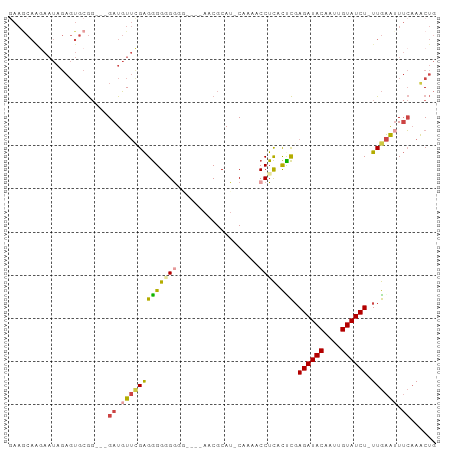
| Location | 26,584,011 – 26,584,105 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.09 |
| Shannon entropy | 0.45068 |
| G+C content | 0.43701 |
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -8.93 |
| Energy contribution | -9.77 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26584011 94 - 27905053 CAGUUUGAAUUUCAA-AGAUACAAUUGUAUCUCGAGUGAGGUUUUG-AUGCGUUUCCCCCCCUCCCCUCGAACAUC---CCGCACUCUAUUCUUGCUUC .(((..((((.....-.(((((....)))))(((((.((((....(-(......))....))))..))))).....---.........))))..))).. ( -19.50, z-score = -2.29, R) >droSim1.chr3R 26223438 90 - 27517382 CAGUUUGAAAUUCAA-AGAUACAAUUGUAUCUCGGGUGAGGUUUUG-AUGCGUU----CCCCACCCCUCGAACAUC---CCGCACUCUAUUCUUGCUUC ..((((((.......-((((((....)))))).(((((.((.....-.......----)).))))).))))))...---..(((.........)))... ( -21.70, z-score = -1.98, R) >droSec1.super_4 5402564 90 - 6179234 CAGUUUGAAAUUCAA-AGAUACAAUUGUAUCUCGGGUGAGGUUUUG-AUGCGUU----CCCCACCCCUCGAACAUC---CCGCACUCUAUUCUUGCUUC ..((((((.......-((((((....)))))).(((((.((.....-.......----)).))))).))))))...---..(((.........)))... ( -21.70, z-score = -1.98, R) >droYak2.chr3R 27504930 92 - 28832112 CAGUUGGAAAUUCAA-AGAUACAAUUGUAUCUAGAGUGAGGUUUUG-CUGCGUU-----UUCCCCCCUCGAACAUCGCAACGCACUCUAUUCUUGCUUC .(((.((((......-((((((....))))))((((((..((....-.((((..-----(((.......)))...)))))).)))))).)))).))).. ( -21.60, z-score = -1.66, R) >droEre2.scaffold_4820 9055061 88 + 10470090 CAGCUUGAAAUUCAA-AGAUACAAUUGUAUCUCGAGUGAGGUUUUG-AUGCGUU------UCCCCCCUCAAACAUC---CCGCACUCUAUUUCUGUUUU .(((..(((((....-((((((....)))))).(((((.((.((((-(.(.(..------....)).)))))....---)).))))).))))).))).. ( -17.50, z-score = -1.84, R) >droAna3.scaffold_12911 963784 83 + 5364042 CUGUUUCCAACCUAAUAGAUACAAUCGUAUCUCAAAUAAGGUUUCGAAUGCGUUCUACUACCCCCUCCCUUAAAUCGCUCUUC---------------- ..((((..(((((.((((((((....))))))...)).)))))..))))(((.......................))).....---------------- ( -11.00, z-score = -2.36, R) >consensus CAGUUUGAAAUUCAA_AGAUACAAUUGUAUCUCGAGUGAGGUUUUG_AUGCGUU____CCCCCCCCCUCGAACAUC___CCGCACUCUAUUCUUGCUUC ..((((((..((((..((((((....))))))....))))((.......))................)))))).......................... ( -8.93 = -9.77 + 0.84)

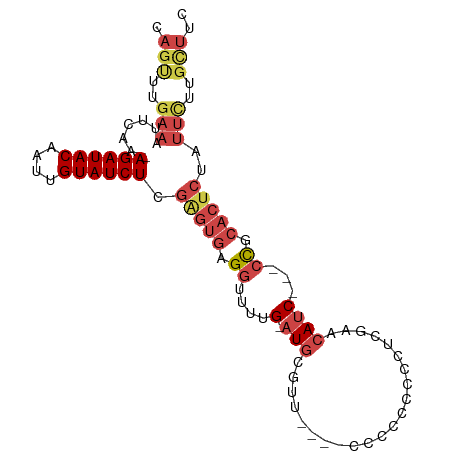
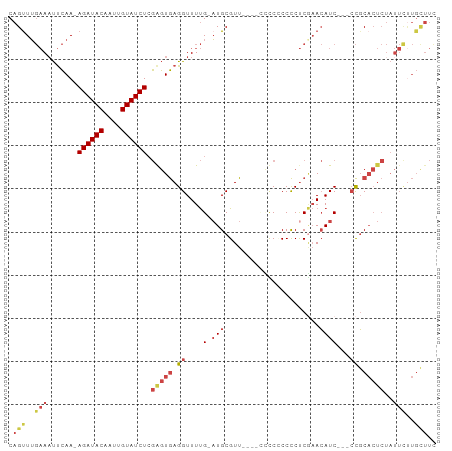
| Location | 26,584,048 – 26,584,144 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 82.66 |
| Shannon entropy | 0.30894 |
| G+C content | 0.41339 |
| Mean single sequence MFE | -20.64 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26584048 96 + 27905053 GGGGAAACGCAUCAAAACCUCACUCGAGAUACAAUUGUAUCUUUGAAAUUCAAACUGAUUUGUAUGCCCUUGCUUUGGGUAGUUCGCUUCAGUUCG- (.(....).).........(((...(((((((....))))))))))......((((((..((..(((((.......)))))...))..))))))..- ( -25.10, z-score = -2.18, R) >droSim1.chr3R 26223475 92 + 27517382 ----GAACGCAUCAAAACCUCACCCGAGAUACAAUUGUAUCUUUGAAUUUCAAACUGAUUUGUAUGCCCUUGCUUUGGGUAGUUCGCCUCACUUUU- ----((((...........(((...(((((((....))))))))))..................(((((.......)))))))))...........- ( -18.60, z-score = -1.80, R) >droSec1.super_4 5402601 92 + 6179234 ----GAACGCAUCAAAACCUCACCCGAGAUACAAUUGUAUCUUUGAAUUUCAAACUGAUUCGUGUGCCCUUACUUUGGGUAGUUUGCCUCACUUUU- ----....(((.........(((..(((((((....))))))).(((((.......))))))))(((((.......)))))...))).........- ( -19.30, z-score = -2.00, R) >droYak2.chr3R 27504969 79 + 28832112 ----AAACGCAGCAAAACCUCACUCUAGAUACAAUUGUAUCUUUGAAUUUCCAACUGAUACGUAUGCCCUUGCUUUAGGUCGC-------------- ----....((.((......(((....((((((....)))))).)))........((((...(((......))).)))))).))-------------- ( -11.10, z-score = -0.24, R) >droEre2.scaffold_4820 9055096 93 - 10470090 ----AAACGCAUCAAAACCUCACUCGAGAUACAAUUGUAUCUUUGAAUUUCAAGCUGAUUCGUAUGCCCUUGCUUUGGGCAGUUCGCAUCAGCCUGG ----...............(((...(((((((....))))))))))....((.((((((.((..(((((.......)))))...)).)))))).)). ( -29.10, z-score = -3.74, R) >consensus ____AAACGCAUCAAAACCUCACUCGAGAUACAAUUGUAUCUUUGAAUUUCAAACUGAUUCGUAUGCCCUUGCUUUGGGUAGUUCGCCUCACUUUG_ ......(((.((((...........(((((((....)))))))............)))).))).(((((.......)))))................ (-14.56 = -14.96 + 0.40)

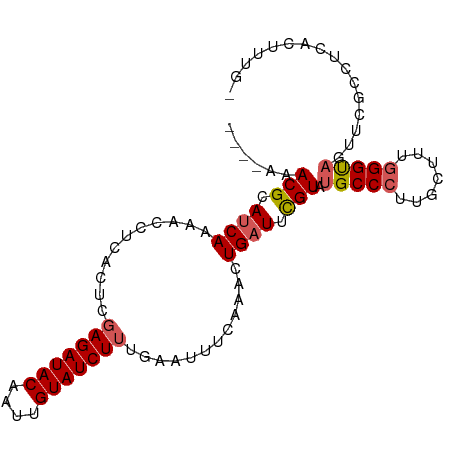
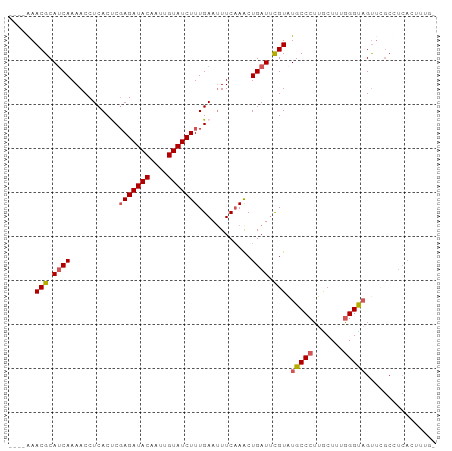
| Location | 26,584,066 – 26,584,181 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 62.12 |
| Shannon entropy | 0.70084 |
| G+C content | 0.40389 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.09 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26584066 115 + 27905053 CUCACUCGAGAUACAAUUGUAUCUUUGAAAUUCAAACUGAUUUGUAUGCCCUUGCUUUGGGUAGUUCGCUUCAGUUCG-GAAUGUUUUCUGUAUAA-UCAUUAGGGAAUAUGGAUGC .(((...(((((((....))))))))))..(((.((((((..((..(((((.......)))))...))..)))))).)-))(((((..(((.....-....)))..)))))...... ( -30.50, z-score = -2.31, R) >droSim1.chr3R 26223489 115 + 27517382 CUCACCCGAGAUACAAUUGUAUCUUUGAAUUUCAAACUGAUUUGUAUGCCCUUGCUUUGGGUAGUUCGCCUCACUUUU-AAUUGUUUUCUGUAUAA-UCACUAGGGAAUAUGGGUGC ..((((((((((((....)))))))............(((..((..(((((.......)))))...))..))).....-...((((..((((....-..)).))..)))).))))). ( -28.00, z-score = -1.77, R) >droSec1.super_4 5402615 115 + 6179234 CUCACCCGAGAUACAAUUGUAUCUUUGAAUUUCAAACUGAUUCGUGUGCCCUUACUUUGGGUAGUUUGCCUCACUUUU-AAUUGUUUUCUGUAUAA-UCAUUAGGGAAUAUGGGUGC ..((((((((((((....))))))).(((((.......)))))((((((((.......)))))........)))....-...((((..(((.....-....)))..)))).))))). ( -28.20, z-score = -1.90, R) >droYak2.chr3R 27504983 88 + 28832112 CUCACUCUAGAUACAAUUGUAUCUUUGAAUUUCCAACUGAUACGUAUGCCCUUGCUUUAGG----UCGCAAAGCAUUAGGGAUCAUGGGUGC------------------------- ..(((((..(((......(((((.(((......)))..))))).....((((((((((...----....))))))..)))))))..))))).------------------------- ( -22.20, z-score = -1.45, R) >droEre2.scaffold_4820 9055110 117 - 10470090 CUCACUCGAGAUACAAUUGUAUCUUUGAAUUUCAAGCUGAUUCGUAUGCCCUUGCUUUGGGCAGUUCGCAUCAGCCUGGAAAUACUUUCUAUAGAAGUCAUCAGCGAUUAUGGGCGC .(((...(((((((....)))))))))).(((((.((((((.((..(((((.......)))))...)).)))))).)))))......(((((((..((.....))..)))))))... ( -35.90, z-score = -2.50, R) >dp4.chr2 30175291 94 - 30794189 UCCAGUCGAGAUACAAUUGUAUCUGUGAAUUGGAUUAUUAUUUGCCAGCCCUUUUAUGGGGUGCGAGCACGAGUAUUCGAGGUUUUCUGAGAAG----------------------- ((((((((((((((....)))))).)).)))))).((((...((((.(((((.....)))))..).)))..))))(((..((....))..))).----------------------- ( -25.60, z-score = -1.55, R) >droPer1.super_7 1042053 94 - 4445127 UCUAGUCGAGAUACAAUUGUAUCUGUGAAUUGGAUUAUUAUUUGCCAGCCCUUUUAUGGGGUGCGAGCACGAGUAUUCGAGGUUUUCUGAGAAG----------------------- ((((((((((((((....)))))).)).)))))).((((...((((.(((((.....)))))..).)))..))))(((..((....))..))).----------------------- ( -23.30, z-score = -0.82, R) >consensus CUCACUCGAGAUACAAUUGUAUCUUUGAAUUUCAAACUGAUUUGUAUGCCCUUGCUUUGGGUAGUUCGCCUCACAUUCGAAAUGUUUUCUGUAGAA_UCA__AG_GA_UAUGG__GC ........((((((....)))))).......................((((.......))))....................................................... ( -9.90 = -10.09 + 0.18)
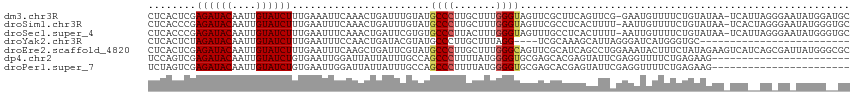
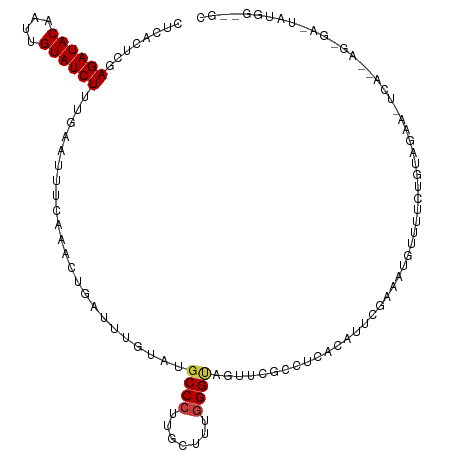
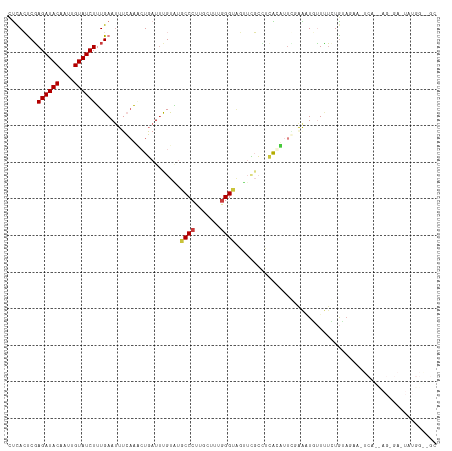
| Location | 26,584,066 – 26,584,181 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 62.12 |
| Shannon entropy | 0.70084 |
| G+C content | 0.40389 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -8.61 |
| Energy contribution | -8.23 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26584066 115 - 27905053 GCAUCCAUAUUCCCUAAUGA-UUAUACAGAAAACAUUC-CGAACUGAAGCGAACUACCCAAAGCAAGGGCAUACAAAUCAGUUUGAAUUUCAAAGAUACAAUUGUAUCUCGAGUGAG ....................-............(((((-((((((((.........(((.......)))........))))))))........((((((....)))))).))))).. ( -21.53, z-score = -1.86, R) >droSim1.chr3R 26223489 115 - 27517382 GCACCCAUAUUCCCUAGUGA-UUAUACAGAAAACAAUU-AAAAGUGAGGCGAACUACCCAAAGCAAGGGCAUACAAAUCAGUUUGAAAUUCAAAGAUACAAUUGUAUCUCGGGUGAG .(((((..............-.................-.....((((.((((((.(((.......)))..........))))))...)))).((((((....)))))).))))).. ( -23.00, z-score = -1.36, R) >droSec1.super_4 5402615 115 - 6179234 GCACCCAUAUUCCCUAAUGA-UUAUACAGAAAACAAUU-AAAAGUGAGGCAAACUACCCAAAGUAAGGGCACACGAAUCAGUUUGAAAUUCAAAGAUACAAUUGUAUCUCGGGUGAG .(((((..............-.................-.....((((.((((((.(((.......)))(....)....))))))...)))).((((((....)))))).))))).. ( -23.60, z-score = -1.83, R) >droYak2.chr3R 27504983 88 - 28832112 -------------------------GCACCCAUGAUCCCUAAUGCUUUGCGA----CCUAAAGCAAGGGCAUACGUAUCAGUUGGAAAUUCAAAGAUACAAUUGUAUCUAGAGUGAG -------------------------.((((((((((((((..(((((((...----..))))))))))).......))))..)))........((((((....))))))...))).. ( -21.21, z-score = -1.77, R) >droEre2.scaffold_4820 9055110 117 + 10470090 GCGCCCAUAAUCGCUGAUGACUUCUAUAGAAAGUAUUUCCAGGCUGAUGCGAACUGCCCAAAGCAAGGGCAUACGAAUCAGCUUGAAAUUCAAAGAUACAAUUGUAUCUCGAGUGAG ..........(((((.................(.(((((.((((((((.((...(((((.......)))))..)).))))))))))))).)..((((((....))))))..))))). ( -34.40, z-score = -2.80, R) >dp4.chr2 30175291 94 + 30794189 -----------------------CUUCUCAGAAAACCUCGAAUACUCGUGCUCGCACCCCAUAAAAGGGCUGGCAAAUAAUAAUCCAAUUCACAGAUACAAUUGUAUCUCGACUGGA -----------------------............((((((.......((((.((.((........)))).))))...................(((((....)))))))))..)). ( -17.80, z-score = -1.25, R) >droPer1.super_7 1042053 94 + 4445127 -----------------------CUUCUCAGAAAACCUCGAAUACUCGUGCUCGCACCCCAUAAAAGGGCUGGCAAAUAAUAAUCCAAUUCACAGAUACAAUUGUAUCUCGACUAGA -----------------------((..((.(((.....(((....)))((((.((.((........)))).)))).............)))..((((((....)))))).))..)). ( -16.80, z-score = -1.41, R) >consensus GC__CCAUA_UC_CU__UGA_UUAUACAGAAAACAUUUCAAAAGUGAGGCGAACUACCCAAAGCAAGGGCAUACAAAUCAGUUUGAAAUUCAAAGAUACAAUUGUAUCUCGAGUGAG ........................................................(((.......)))...................((((.((((((....))))))....)))) ( -8.61 = -8.23 + -0.38)
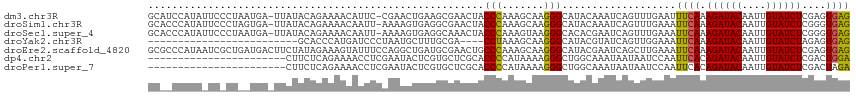
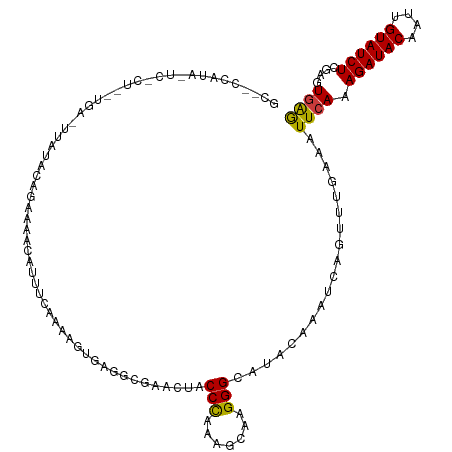
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:10 2011