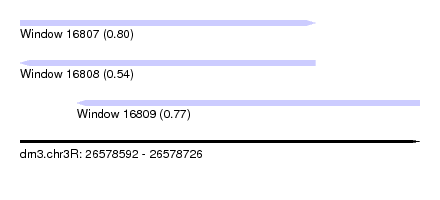
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,578,592 – 26,578,726 |
| Length | 134 |
| Max. P | 0.799010 |

| Location | 26,578,592 – 26,578,691 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.92 |
| Shannon entropy | 0.29833 |
| G+C content | 0.38965 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.34 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
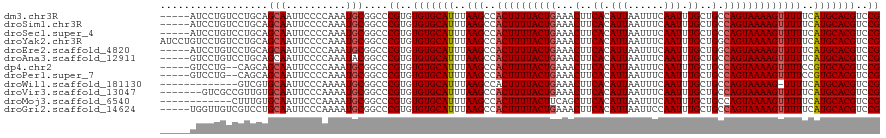
>dm3.chr3R 26578592 99 + 27905053 ---------------------AUCCCCAAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ---------------------.(((.((((......)))).))).((((((.....((((((((((((..((.((((....)))).))..))..)))))))))).......))))))... ( -25.90, z-score = -2.15, R) >droSim1.chr3R 26217996 99 + 27517382 ---------------------AUCCCCAAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ---------------------.(((.((((......)))).))).((((((.....((((((((((((..((.((((....)))).))..))..)))))))))).......))))))... ( -25.90, z-score = -2.15, R) >droSec1.super_4 5397129 99 + 6179234 ---------------------AUCCCCAAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ---------------------.(((.((((......)))).))).((((((.....((((((((((((..((.((((....)))).))..))..)))))))))).......))))))... ( -25.90, z-score = -2.15, R) >droYak2.chr3R 27499327 99 + 28832112 ---------------------AUCCCCAAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGCCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ---------------------.(((.((((......)))).))).((((((.....((((((((((....((.((((....)))).))......)))))))))).......))))))... ( -24.70, z-score = -2.13, R) >droEre2.scaffold_4820 9049633 99 - 10470090 ---------------------AUCCCCAAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGCCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ---------------------.(((.((((......)))).))).((((((.....((((((((((....((.((((....)))).))......)))))))))).......))))))... ( -24.70, z-score = -2.13, R) >droAna3.scaffold_12911 958287 103 - 5364042 -----------------CCGAGGCCCCAAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA -----------------(((..((........))......)))..((((((.....((((((((((((..((.((((....)))).))..))..)))))))))).......))))))... ( -25.10, z-score = -1.01, R) >dp4.chr2 30168293 116 - 30794189 ----GGAGACUCCCCGUUUGAGGCCCCAAAAAGCAAUUUGCGGACGUGCACGGAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ----(....)...((((.....((........)).....))))..(((((.((...((((((((((((..((.((((....)))).))..))..))))))))))..))....)))))... ( -28.00, z-score = -0.17, R) >droPer1.super_7 1034779 116 - 4445127 ----GGAGACUCCCCGUUUGAGGCCCCAAAAAGCAAUUUGCGGACGUGCACGGAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ----(....)...((((.....((........)).....))))..(((((.((...((((((((((((..((.((((....)))).))..))..))))))))))..))....)))))... ( -28.00, z-score = -0.17, R) >droWil1.scaffold_181130 10109788 115 + 16660200 ---CGAGAACUCACUUCGAAAAGACCC-AAAAGCAAUUUGCGGACGUGCAUG-AAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ---((((.......))))......(((-(((.....)))).))..((((((.-...((((((((((((..((.((((....)))).))..))..)))))))))).......))))))... ( -25.30, z-score = -0.90, R) >droVir3.scaffold_13047 11377573 117 + 19223366 ---GGGACACACACGUCUGUUAGUCCCCAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ---.((((......))))....((((.((((.....)))).))))((((((.....((((((((((((..((.((((....)))).))..))..)))))))))).......))))))... ( -32.90, z-score = -2.03, R) >droMoj3.scaffold_6540 3782105 120 - 34148556 ACGGGCACACACACGUCUUUGAGUCCCCAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGCUGAAGUAAAAGUGGCUUAAAUGCACACA ..((((........))))....((((.((((.....)))).))))((((((.....(((((((((..((((..((((....)))).....)))).))))))))).......))))))... ( -33.20, z-score = -1.60, R) >droGri2.scaffold_14624 1409727 116 + 4233967 ---CACACACACACGUAUGU-AGUCCCCAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGGAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ---.(((.((....)).)))-.((((.((((.....)))).))))((((((.....((((((((((((..((.((((....)))).))..))..)))))))))).......))))))... ( -30.40, z-score = -1.52, R) >consensus _____________________AGCCCCAAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACA ................................((.....))....((((((.....((((((((((((..((.((((....)))).))..))..)))))))))).......))))))... (-21.99 = -22.34 + 0.35)


| Location | 26,578,592 – 26,578,691 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Shannon entropy | 0.29833 |
| G+C content | 0.38965 |
| Mean single sequence MFE | -27.19 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.44 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26578592 99 - 27905053 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUUGGGGAU--------------------- ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))((((.((((......)))).))))--------------------- ( -25.60, z-score = -2.29, R) >droSim1.chr3R 26217996 99 - 27517382 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUUGGGGAU--------------------- ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))((((.((((......)))).))))--------------------- ( -25.60, z-score = -2.29, R) >droSec1.super_4 5397129 99 - 6179234 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUUGGGGAU--------------------- ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))((((.((((......)))).))))--------------------- ( -25.60, z-score = -2.29, R) >droYak2.chr3R 27499327 99 - 28832112 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGGCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUUGGGGAU--------------------- ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))((((.((((......)))).))))--------------------- ( -27.80, z-score = -2.81, R) >droEre2.scaffold_4820 9049633 99 + 10470090 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGGCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUUGGGGAU--------------------- ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))((((.((((......)))).))))--------------------- ( -27.80, z-score = -2.81, R) >droAna3.scaffold_12911 958287 103 + 5364042 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUUGGGGCCUCGG----------------- ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))((((.((((......)))))))).....----------------- ( -24.90, z-score = -1.52, R) >dp4.chr2 30168293 116 + 30794189 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUCCGUGCACGUCCGCAAAUUGCUUUUUGGGGCCUCAAACGGGGAGUCUCC---- ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))..((.((((......)))).))((((....)))).......---- ( -26.30, z-score = -0.10, R) >droPer1.super_7 1034779 116 + 4445127 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUCCGUGCACGUCCGCAAAUUGCUUUUUGGGGCCUCAAACGGGGAGUCUCC---- ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))..((.((((......)))).))((((....)))).......---- ( -26.30, z-score = -0.10, R) >droWil1.scaffold_181130 10109788 115 - 16660200 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUU-CAUGCACGUCCGCAAAUUGCUUUU-GGGUCUUUUCGAAGUGAGUUCUCG--- ...((((((..((...((((((((((...(..((.(((......))).))..).)))))))))))).-.))))))(.((.((((.....)))-))).)...................--- ( -20.80, z-score = 0.36, R) >droVir3.scaffold_13047 11377573 117 - 19223366 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUGGGGACUAACAGACGUGUGUGUCCC--- (((((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))((((.((((.....)))).))))..)))((((....))))..--- ( -30.20, z-score = -1.68, R) >droMoj3.scaffold_6540 3782105 120 + 34148556 UGUGUGCAUUUAAGCCACUUUUACUUCAGCUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUGGGGACUCAAAGACGUGUGUGUGCCCGU ...((((((..(((..(((((((((.((((...................))))..))))))))))))..))))))((((.((((.....)))).))))......(((.((....)).))) ( -32.51, z-score = -1.60, R) >droGri2.scaffold_14624 1409727 116 - 4233967 UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUCCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUGGGGACU-ACAUACGUGUGUGUGUG--- ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))((((.((((.....)))).))))(-((((((....)))))))--- ( -32.90, z-score = -2.31, R) >consensus UGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUUGGGGCU_____________________ ...((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..))))))....((.....))................................ (-18.50 = -18.44 + -0.05)

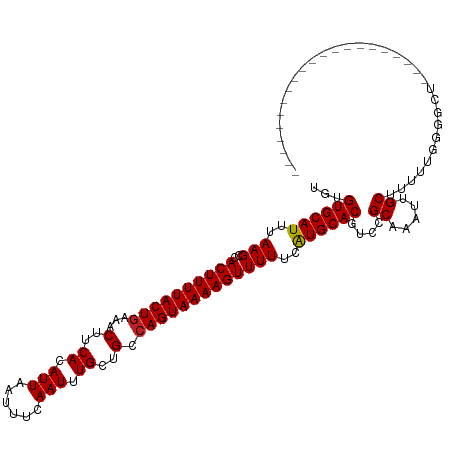
| Location | 26,578,611 – 26,578,726 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.66 |
| Shannon entropy | 0.17975 |
| G+C content | 0.43713 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.86 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26578611 115 - 27905053 -----AUCCUGUCCUGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCG -----.....(..(((((.............)))))..)(..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..). ( -24.62, z-score = -1.99, R) >droSim1.chr3R 26218015 115 - 27517382 -----AUCCUGUCCUGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCG -----.....(..(((((.............)))))..)(..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..). ( -24.62, z-score = -1.99, R) >droSec1.super_4 5397148 115 - 6179234 -----AUCCUGUCCUGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCG -----.....(..(((((.............)))))..)(..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..). ( -24.62, z-score = -1.99, R) >droYak2.chr3R 27499346 120 - 28832112 AUCCUGUCCUGUCCUGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGGCAGUAAAAGUUUUUCAUGCACGUCCG ..........(..(((((.............)))))..)(..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..). ( -26.82, z-score = -1.77, R) >droEre2.scaffold_4820 9049652 115 + 10470090 -----AUCCUGUCCUGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGGCAGUAAAAGUUUUUCAUGCACGUCCG -----.....(..(((((.............)))))..)(..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..). ( -26.82, z-score = -1.96, R) >droAna3.scaffold_12911 958310 115 + 5364042 -----GUCCUGUCCUGCAGCAAUUCCCCAAAUACGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCG -----...(((.....))).......((......))..((..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..)) ( -21.90, z-score = -1.41, R) >dp4.chr2 30168329 113 + 30794189 -----GUCCUG--CAGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUCCGUGCACGUCCG -----(..(((--((................)))))..)(..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..). ( -24.09, z-score = -1.23, R) >droPer1.super_7 1034815 113 + 4445127 -----GUCCUG--CAGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUCCGUGCACGUCCG -----(..(((--((................)))))..)(..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..). ( -24.09, z-score = -1.23, R) >droWil1.scaffold_181130 10109824 106 - 16660200 -------------GUCGUGCAAUUCCCAAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAG-UUUUCAUGCACGUCCG -------------(.((((((.............(((..(((....)))....)))((((((((((...(..((.(((......))).))..).)))))))))-).....)))))).).. ( -23.70, z-score = -1.73, R) >droVir3.scaffold_13047 11377610 113 - 19223366 -------GUCGCCGUUGUGCAAUUCCCAAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCG -------...(((((..((.......))....))))).((..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..)) ( -27.30, z-score = -2.17, R) >droMoj3.scaffold_6540 3782145 108 + 34148556 ------------CUUUGUGCAAUUCCCAAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUUCAGCUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCG ------------....(((((.............(((..(((....)))....)))(((((((((.((((...................))))..)))))))))......)))))..... ( -24.51, z-score = -2.22, R) >droGri2.scaffold_14624 1409763 115 - 4233967 -----UGGUUGUCGUCCUGCAAUUCCCAAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUCCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCG -----.(((((.(((..((.......))..))))))))((..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..)) ( -25.90, z-score = -1.53, R) >consensus _____AUCCUGUCCUGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCG ..................(((..........)))....((..(((((((..(((..((((((((((...(..((.(((......))).))..).)))))))))))))..)))))))..)) (-20.75 = -20.86 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:06 2011