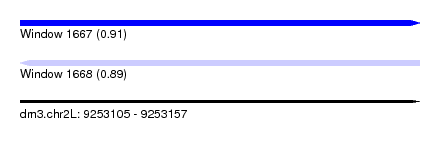
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,253,105 – 9,253,157 |
| Length | 52 |
| Max. P | 0.914116 |

| Location | 9,253,105 – 9,253,157 |
|---|---|
| Length | 52 |
| Sequences | 6 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 67.23 |
| Shannon entropy | 0.61586 |
| G+C content | 0.51582 |
| Mean single sequence MFE | -13.93 |
| Consensus MFE | -7.71 |
| Energy contribution | -8.18 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
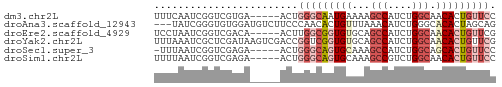
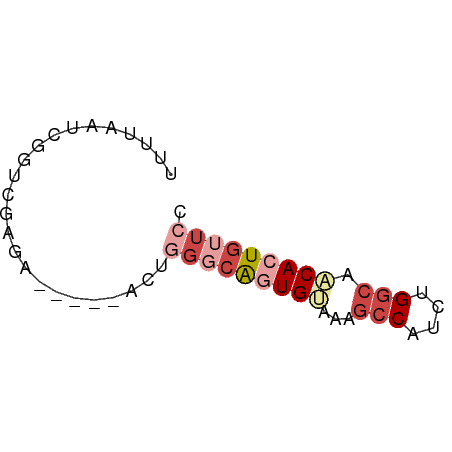
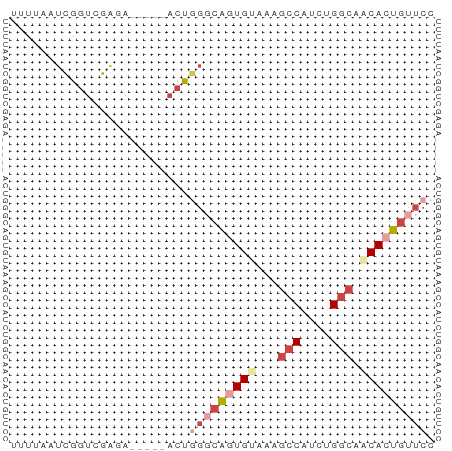
>dm3.chr2L 9253105 52 + 23011544 GGAACAGUGUUGCCAGAUGGCUUUUCAUUGCCCAGU-----UCACGACCGAUUGAAA ((..(((((..(((....)))....))))).))..(-----(((........)))). ( -10.70, z-score = -0.25, R) >droAna3.scaffold_12943 593231 54 + 5039921 CUGCUAGUGUGCCCAGAUGUUUAAACAGUGUUGGGAAGACAUCCACACCCGAUA--- ......((((((((((.(((....)))...)))))........)))))......--- ( -11.30, z-score = -0.28, R) >droEre2.scaffold_4929 9858507 52 + 26641161 CGAACAGUGUUGCCAGAUGGCUGCACACCGCCAAGU-----UGUCGACCGAUUAGGA ......((((.(((....))).))))....((.(((-----((.....))))).)). ( -14.10, z-score = -0.80, R) >droYak2.chr2L 11920744 57 + 22324452 CGAACAGUGUUGCCAGAUGGCUGCACACCGACCGGUCGACUUAUCGAGCGAUUUAAA ......((((.(((....))).))))......((.((((....)))).))....... ( -17.00, z-score = -1.84, R) >droSec1.super_3 4712072 51 + 7220098 GGAACAGUGCUGCCAGAUGGCUUUGCACUGCCCAGU-----UCUCGACCGAUUAAA- ((..((((((.(((....)))...)))))).))...-----...............- ( -16.20, z-score = -2.05, R) >droSim1.chr2L 9029164 52 + 22036055 GGAACAGUGUUGCCAGACGGCUUUGCACUGCCCAGU-----UCUCGACCGAUUAAAA ((..((((((.(((....)))...)))))).))...-----................ ( -14.30, z-score = -1.38, R) >consensus CGAACAGUGUUGCCAGAUGGCUUUACACUGCCCAGU_____UCUCGACCGAUUAAAA ....((((((.(((....)))...))))))........................... ( -7.71 = -8.18 + 0.47)

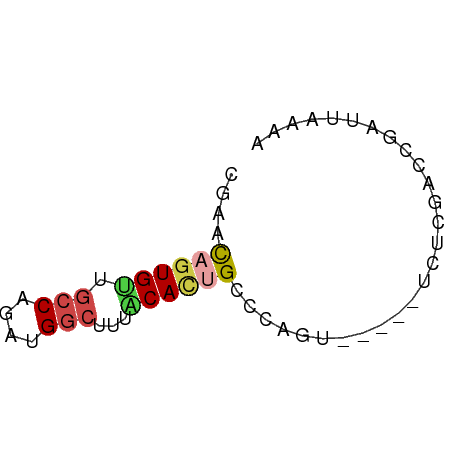
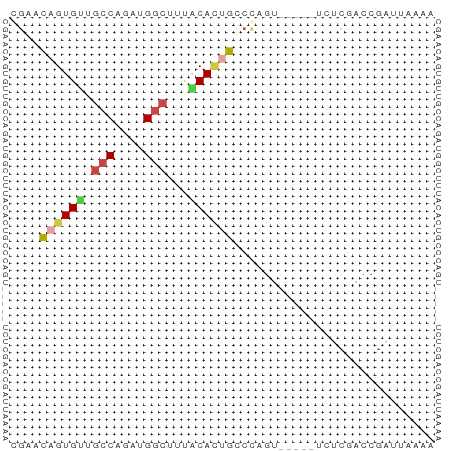
| Location | 9,253,105 – 9,253,157 |
|---|---|
| Length | 52 |
| Sequences | 6 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 67.23 |
| Shannon entropy | 0.61586 |
| G+C content | 0.51582 |
| Mean single sequence MFE | -13.48 |
| Consensus MFE | -8.64 |
| Energy contribution | -10.25 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9253105 52 - 23011544 UUUCAAUCGGUCGUGA-----ACUGGGCAAUGAAAAGCCAUCUGGCAACACUGUUCC (((((.(((((.....-----)))))....))))).(((....)))........... ( -9.60, z-score = 0.15, R) >droAna3.scaffold_12943 593231 54 - 5039921 ---UAUCGGGUGUGGAUGUCUUCCCAACACUGUUUAAACAUCUGGGCACACUAGCAG ---.....((((((........((((....(((....)))..))))))))))..... ( -9.90, z-score = 0.49, R) >droEre2.scaffold_4929 9858507 52 - 26641161 UCCUAAUCGGUCGACA-----ACUUGGCGGUGUGCAGCCAUCUGGCAACACUGUUCG ................-----....((((((((...(((....))).)))))))).. ( -13.40, z-score = -0.54, R) >droYak2.chr2L 11920744 57 - 22324452 UUUAAAUCGCUCGAUAAGUCGACCGGUCGGUGUGCAGCCAUCUGGCAACACUGUUCG .....((((.((((....)))).))))((((((...(((....))).)))))).... ( -17.60, z-score = -1.96, R) >droSec1.super_3 4712072 51 - 7220098 -UUUAAUCGGUCGAGA-----ACUGGGCAGUGCAAAGCCAUCUGGCAGCACUGUUCC -...............-----...(((((((((...(((....))).))))))))). ( -17.40, z-score = -2.07, R) >droSim1.chr2L 9029164 52 - 22036055 UUUUAAUCGGUCGAGA-----ACUGGGCAGUGCAAAGCCGUCUGGCAACACUGUUCC ................-----...((((((((....(((....)))..)))))))). ( -13.00, z-score = -0.47, R) >consensus UUUUAAUCGGUCGAGA_____ACUGGGCAGUGUAAAGCCAUCUGGCAACACUGUUCC ........................(((((((((...(((....))).))))))))). ( -8.64 = -10.25 + 1.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:52 2011