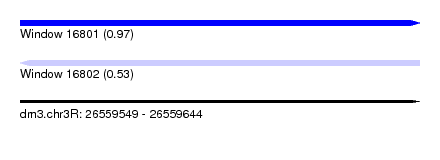
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,559,549 – 26,559,644 |
| Length | 95 |
| Max. P | 0.965749 |

| Location | 26,559,549 – 26,559,644 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 90.94 |
| Shannon entropy | 0.15591 |
| G+C content | 0.45830 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -25.22 |
| Energy contribution | -24.66 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.43 |
| Structure conservation index | 1.03 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
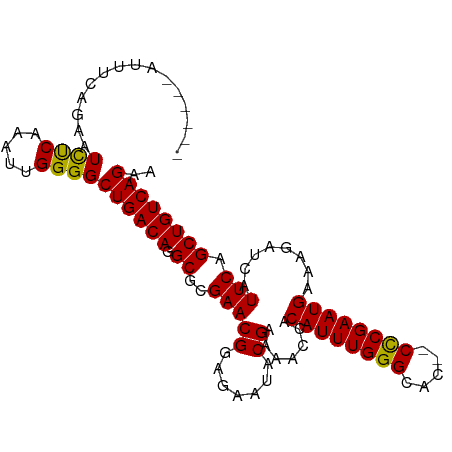
>dm3.chr3R 26559549 95 + 27905053 ------AUUUCAGAAUCUCAAAUUGGGGCUGACAGGCGCGAACGGAGAAUACGAAAACCACAUUUGGGCAC--CCCGAAUGAAAGAUCAUUCAGCUGUCAGAA ------.........((((.....))))(((((((.(..(((((.......)).......((((((((...--))))))))........))).)))))))).. ( -23.70, z-score = -0.90, R) >droAna3.scaffold_12911 938445 92 - 5364042 -----------CGAAUUUCAAAUUGGGGCUGACAAGCGCGAACGGAGAAUACGAAAAACACAUUUGGGCACAACUCGAAUGAAAGAUCAUUCAGCUGUCAGAA -----------.................((((((.((..(((((.......)).......((((((((.....))))))))........))).)))))))).. ( -20.30, z-score = -1.46, R) >droYak2.chr3R 27479542 101 + 28832112 UUUAGUAUUUCAGAAUCUCAAAUUGGGGCUGACAGGCGCGAACGGAGUAUACGAAAACCACAUUUGGGCAA--CCCGAAUGAAAGAUCAUUCAGCUGUCAGAA ...............((((.....))))(((((((((.((((.(..((........))..).)))).))..--...((((((....))))))..))))))).. ( -25.50, z-score = -1.45, R) >droSec1.super_4 5378157 95 + 6179234 ------AUUUCAGAAUCCCAAAUUGGGGCUGACAGGCGCGAACGGAGAAUACGAAAACCACAUUUGGGCAC--CCCGAAUGAAAGAUCAUUCAGCUGUCAGAA ------.........((((.....))))(((((((.(..(((((.......)).......((((((((...--))))))))........))).)))))))).. ( -26.70, z-score = -1.77, R) >droSim1.chr3R 26198973 95 + 27517382 ------AUCUCAGAAUCCCAAAUUGGGGCUGACAGGCGCGAACGGAGAAUACGAAAACCACAUUUGGGCAC--CCCGAAUGAAAGAUCAUUCAGCUGUCAGAA ------.........((((.....))))(((((((.(..(((((.......)).......((((((((...--))))))))........))).)))))))).. ( -26.70, z-score = -1.55, R) >consensus ______AUUUCAGAAUCUCAAAUUGGGGCUGACAGGCGCGAACGGAGAAUACGAAAACCACAUUUGGGCAC__CCCGAAUGAAAGAUCAUUCAGCUGUCAGAA ...............((((.....))))((((((.((..(((((.......)).......((((((((.....))))))))........))).)))))))).. (-25.22 = -24.66 + -0.56)

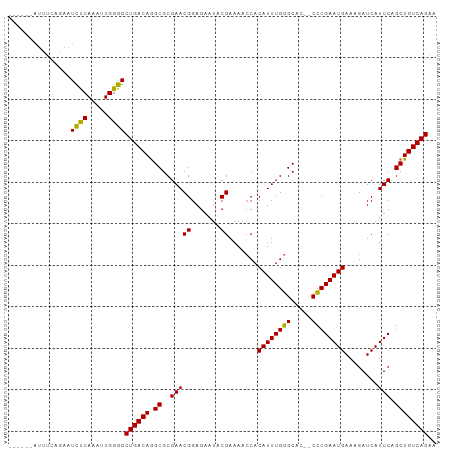
| Location | 26,559,549 – 26,559,644 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 90.94 |
| Shannon entropy | 0.15591 |
| G+C content | 0.45830 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
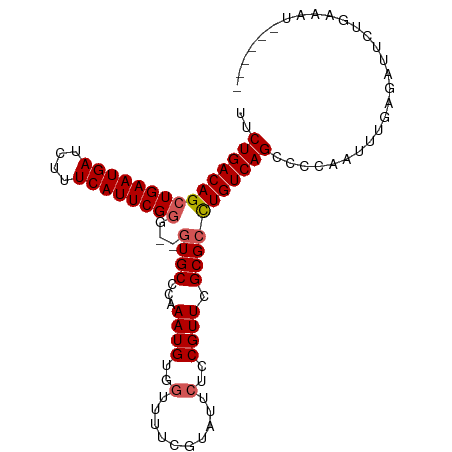
>dm3.chr3R 26559549 95 - 27905053 UUCUGACAGCUGAAUGAUCUUUCAUUCGGG--GUGCCCAAAUGUGGUUUUCGUAUUCUCCGUUCGCGCCUGUCAGCCCCAAUUUGAGAUUCUGAAAU------ ..((((((.((((((((....))))))))(--((((...((((..(..(....)..)..)))).)))))))))))......................------ ( -25.50, z-score = -1.36, R) >droAna3.scaffold_12911 938445 92 + 5364042 UUCUGACAGCUGAAUGAUCUUUCAUUCGAGUUGUGCCCAAAUGUGUUUUUCGUAUUCUCCGUUCGCGCUUGUCAGCCCCAAUUUGAAAUUCG----------- ..((((((((.((((((....))))))(((..((((..(((.....)))..)))).))).......)).)))))).................----------- ( -19.70, z-score = -1.86, R) >droYak2.chr3R 27479542 101 - 28832112 UUCUGACAGCUGAAUGAUCUUUCAUUCGGG--UUGCCCAAAUGUGGUUUUCGUAUACUCCGUUCGCGCCUGUCAGCCCCAAUUUGAGAUUCUGAAAUACUAAA ..(((((((((((((((....)))))))).--..((.(.((((..((........))..)))).).))))))))).....((((.((...)).))))...... ( -24.50, z-score = -1.62, R) >droSec1.super_4 5378157 95 - 6179234 UUCUGACAGCUGAAUGAUCUUUCAUUCGGG--GUGCCCAAAUGUGGUUUUCGUAUUCUCCGUUCGCGCCUGUCAGCCCCAAUUUGGGAUUCUGAAAU------ ..((((((.((((((((....))))))))(--((((...((((..(..(....)..)..)))).))))))))))).(((.....)))..........------ ( -29.70, z-score = -2.20, R) >droSim1.chr3R 26198973 95 - 27517382 UUCUGACAGCUGAAUGAUCUUUCAUUCGGG--GUGCCCAAAUGUGGUUUUCGUAUUCUCCGUUCGCGCCUGUCAGCCCCAAUUUGGGAUUCUGAGAU------ ..((((((.((((((((....))))))))(--((((...((((..(..(....)..)..)))).))))))))))).(((.....)))..........------ ( -29.70, z-score = -1.73, R) >consensus UUCUGACAGCUGAAUGAUCUUUCAUUCGGG__GUGCCCAAAUGUGGUUUUCGUAUUCUCCGUUCGCGCCUGUCAGCCCCAAUUUGAGAUUCUGAAAU______ ..(((((((((((((((....))))))))...((((...((((..(..........)..)))).)))))))))))............................ (-20.42 = -20.86 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:00 2011