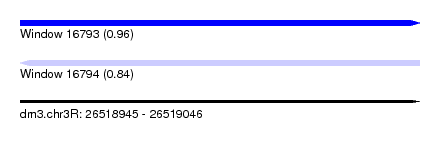
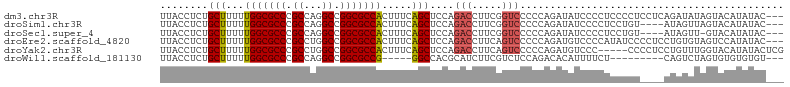
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,518,945 – 26,519,046 |
| Length | 101 |
| Max. P | 0.956986 |

| Location | 26,518,945 – 26,519,046 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.29 |
| Shannon entropy | 0.42229 |
| G+C content | 0.57663 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -27.15 |
| Energy contribution | -27.57 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.956986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
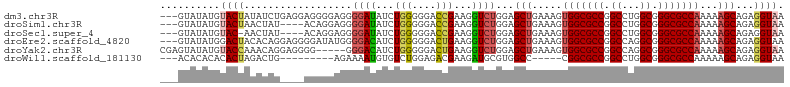
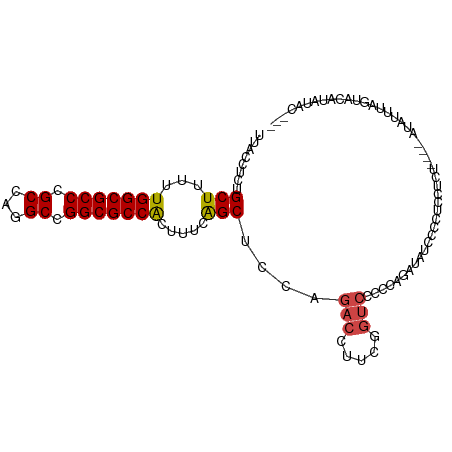
>dm3.chr3R 26518945 101 + 27905053 ---GUAUAUGUACUAUAUCUGAGGAGGGGAGGGGAUAUCUGGGGGACCGAAGGUCUGGAGCUGAAAGUGGCGCCGGCCUGGCGGGCGCCAAAAAGCAGAGGUAA ---............(((((............((((...(((....)))...))))...(((.....(((((((.((...)).)))))))...)))..))))). ( -33.50, z-score = -1.13, R) >droSim1.chr3R 26158485 97 + 27517382 ---GUAUAUGUACUAACUAU----ACAGGAGGGGAUAUCUGGGGGACCGAAGGUCUGGAGCUGAAAGUGGCGCCGGCCUGGCGGGCGCCAAAAAGCAGAGGUAA ---(((((.((....)))))----))......((((...(((....)))...))))...(((.....(((((((.((...)).)))))))...)))........ ( -33.60, z-score = -1.65, R) >droSec1.super_4 5337873 96 + 6179234 ---GUAUAUGUAC-AACUAU----ACAGGAGGGGAUAUCUGGGGGACCGAAGGUCUGGAGCUGAAAGUGGCGCCGGCCUGGCGGGCGCCAAAAAGCAGAGGUAA ---(((((.....-...)))----))......((((...(((....)))...))))...(((.....(((((((.((...)).)))))))...)))........ ( -32.90, z-score = -1.67, R) >droEre2.scaffold_4820 8984314 101 - 10470090 ---GUAUAUGGACUACACAGGAGGGGAUAUGGGGACAUCUGGGGGACUGAAGGUCUGGAGCUGAAAGUGGCGCCGGCCAGGCGGGCGCCAAAAAGCAGAGGUAA ---......(((((...(((........(((....)))........)))..)))))...(((.....(((((((.((...)).)))))))...)))........ ( -32.69, z-score = -1.78, R) >droYak2.chr3R 27437422 99 + 28832112 CGAGUAUAUGUACCAAACAGGAGGGG-----GGGACAUCUGGGGGACUGAAGGUCUGGAGCUGAAAGUGGCGCCGGCCAGGCGGGCGCCAAAAAGCAGAGGUAA ..........((((...((((((..(-----....)..))((....)).....))))..(((.....(((((((.((...)).)))))))...)))...)))). ( -35.30, z-score = -2.40, R) >droWil1.scaffold_181130 10032458 87 + 16660200 ---ACACACACACUAGACUG---------AGAAAAUGUGUCUGGAGACGAAGAUGCGUGGCC-----CGGCGCCGGCCUGGCGGGCGCCAAAAAGCAGAGGUAA ---..............(((---------.....(((((((((....)..))))))))....-----.((((((.((...)).))))))......)))...... ( -27.30, z-score = -0.26, R) >consensus ___GUAUAUGUACUAAACAG____AGAGGAGGGGAUAUCUGGGGGACCGAAGGUCUGGAGCUGAAAGUGGCGCCGGCCUGGCGGGCGCCAAAAAGCAGAGGUAA ..........((((..................((((...(((....)))...))))...(((.....(((((((.((...)).)))))))...)))...)))). (-27.15 = -27.57 + 0.42)
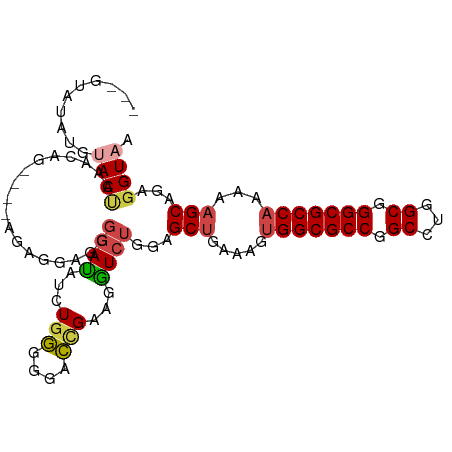
| Location | 26,518,945 – 26,519,046 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.29 |
| Shannon entropy | 0.42229 |
| G+C content | 0.57663 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -17.79 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.841602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26518945 101 - 27905053 UUACCUCUGCUUUUUGGCGCCCGCCAGGCCGGCGCCACUUUCAGCUCCAGACCUUCGGUCCCCCAGAUAUCCCCUCCCCUCCUCAGAUAUAGUACAUAUAC--- .....((((.....(((((((.((...)).)))))))............((((...))))...))))..................................--- ( -21.50, z-score = -1.00, R) >droSim1.chr3R 26158485 97 - 27517382 UUACCUCUGCUUUUUGGCGCCCGCCAGGCCGGCGCCACUUUCAGCUCCAGACCUUCGGUCCCCCAGAUAUCCCCUCCUGU----AUAGUUAGUACAUAUAC--- .....((((.....(((((((.((...)).)))))))............((((...))))...))))..........(((----((.....))))).....--- ( -23.60, z-score = -1.46, R) >droSec1.super_4 5337873 96 - 6179234 UUACCUCUGCUUUUUGGCGCCCGCCAGGCCGGCGCCACUUUCAGCUCCAGACCUUCGGUCCCCCAGAUAUCCCCUCCUGU----AUAGUU-GUACAUAUAC--- .....((((.....(((((((.((...)).)))))))............((((...))))...))))..........(((----((....-))))).....--- ( -23.60, z-score = -1.54, R) >droEre2.scaffold_4820 8984314 101 + 10470090 UUACCUCUGCUUUUUGGCGCCCGCCUGGCCGGCGCCACUUUCAGCUCCAGACCUUCAGUCCCCCAGAUGUCCCCAUAUCCCCUCCUGUGUAGUCCAUAUAC--- ......((((....(((((((.((...)).)))))))....(((.....(((.....))).....(((((....))))).....))).)))).........--- ( -21.70, z-score = -1.30, R) >droYak2.chr3R 27437422 99 - 28832112 UUACCUCUGCUUUUUGGCGCCCGCCUGGCCGGCGCCACUUUCAGCUCCAGACCUUCAGUCCCCCAGAUGUCCC-----CCCCUCCUGUUUGGUACAUAUACUCG .((((...(((...(((((((.((...)).))))))).....)))....(((.....))).............-----............)))).......... ( -21.80, z-score = -0.97, R) >droWil1.scaffold_181130 10032458 87 - 16660200 UUACCUCUGCUUUUUGGCGCCCGCCAGGCCGGCGCCG-----GGCCACGCAUCUUCGUCUCCAGACACAUUUUCU---------CAGUCUAGUGUGUGUGU--- ........(((...(((((((.((...)).)))))))-----)))(((((......(((....)))(((((....---------......)))))))))).--- ( -26.60, z-score = -0.73, R) >consensus UUACCUCUGCUUUUUGGCGCCCGCCAGGCCGGCGCCACUUUCAGCUCCAGACCUUCGGUCCCCCAGAUAUCCCCUCCUCU____AUAUUUAGUACAUAUAC___ ........(((...(((((((.((...)).))))))).....)))....(((.....)))............................................ (-17.79 = -18.02 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:54 2011