
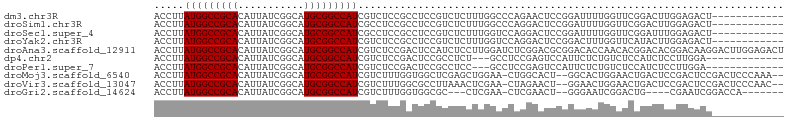
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,501,835 – 26,501,928 |
| Length | 93 |
| Max. P | 0.999221 |

| Location | 26,501,835 – 26,501,928 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 66.70 |
| Shannon entropy | 0.67278 |
| G+C content | 0.57506 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -14.40 |
| Energy contribution | -14.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26501835 93 + 27905053 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGUCUCCGCCUCCGUCUCUUUGGCCCAGAACUCCGGAUUUUGGUUCGGACUUGGAGACU------------ .....(((((((((...........))))))))).(((((((((...........))).......(((((((....)))))))...)))))).------------ ( -36.00, z-score = -2.90, R) >droSim1.chr3R 26148935 93 + 27517382 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGCCUCCGCCUCCGUCUCUUUGGCCCAGGACUCCGGAUUUUGGUUCGGACUUGGAGACU------------ .....(((((((((...........)))))))))...((((.(((..(((.....)))..)))..(((((((....)))))))...))))...------------ ( -33.00, z-score = -1.32, R) >droSec1.super_4 5328326 93 + 6179234 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGCCUCCGCCUCCGUCUCUUUGGUCCAGGACUCCGGAUUUUGGUUCGGAUUUGGAGACU------------ .....(((((((((...........)))))))))...(((((..((((...((..(((((.((...)))))))..))..))))..)))))...------------ ( -34.80, z-score = -2.01, R) >droYak2.chr3R 27427539 93 + 28832112 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGUCUCCGCCUCCGUCUCUUUGGUCCAGGACUCCGGACUUUGGUUCAUACUUGGAGACU------------ .....(((((((((...........))))))))).(((((((.........((..(((((.((...)))))))..))........))))))).------------ ( -33.33, z-score = -2.67, R) >droAna3.scaffold_12911 882804 105 - 5364042 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGUCUCCGACUCCAUCUCCUUGGAUCUCGGACGCGGACACCAACACGGACACGGACAAGGACUUGGAGACU .....(((((((((...........))))))))).((((((((.(((...((....))....(..((.(..(........)..).))..)..))).)))))))). ( -38.10, z-score = -3.25, R) >dp4.chr2 30093606 89 - 30794189 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGUCUCCGACUCCGCCUCU---GCCUCCGAGUCCAUUCUCUGUCUCCAUCUCCUUGGA------------- .....(((((((((...........))))))))).......(((((.((....---))....)))))...........((((......))))------------- ( -23.00, z-score = -1.73, R) >droPer1.super_7 959576 89 - 4445127 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGUCUCCGACUCCGCCUCC---GCCUCCGAGUCCAUUCUCUGUCUCCAUCUCCUUGGA------------- .....(((((((((...........))))))))).......(((((.((....---))....)))))...........((((......))))------------- ( -23.10, z-score = -1.73, R) >droMoj3.scaffold_6540 3692724 100 - 34148556 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGUCUUUGGUGGCUCGAGCUGGAA-CUGGCACU--GGCACUGGAACUGACUCCGACUCCGACUCCCAAA-- .....(((((((((...........))))))))).....((((..(.(((.((..(..-......).--.))..((((......))))....))))..)))).-- ( -30.40, z-score = -0.55, R) >droVir3.scaffold_13047 11295268 100 + 19223366 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGUCUUUGGCGCCUUAAACUCGAA-CUAGAACU--GGAACUGGAACUGACUCCGACUCCGACUCCCAAC-- .....(((((((((...........))))))))).((((((((..(.........)..-)))))...--(((..((((......))))..)))))).......-- ( -24.50, z-score = -1.44, R) >droGri2.scaffold_14624 1294876 88 + 4233967 ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGUCUUUGGUGGCGC---CUCGAA-CUCGAACU--GGGAAUCGGACUG----CGAAUCGGACCA------- .((...((((((((...........))))))))((((.((((((..(.(---((((..-..)))...--))).))))))..)----)))...))....------- ( -25.20, z-score = 0.65, R) >consensus ACCUUAUGGCCGCACAUUAUCGGCAUGCGGCCAUCGUCUCCGCCUCCGUCUCCUUGGACCAGGAAUCCGGACUCUGGUUCGGACUCCGAGACU____________ .....(((((((((...........)))))))))....................................................................... (-14.40 = -14.40 + 0.00)
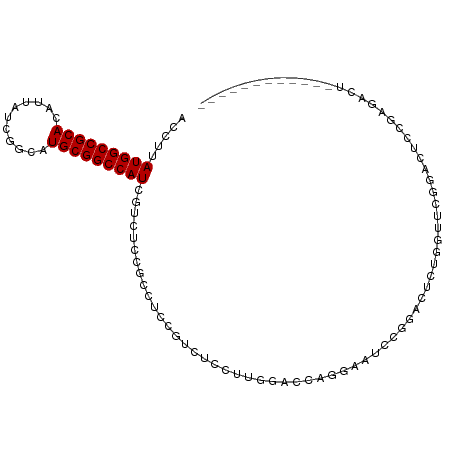
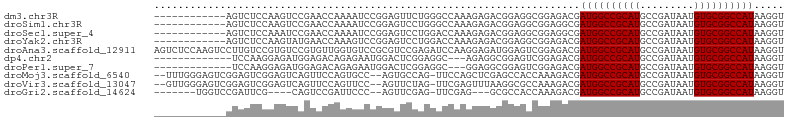
| Location | 26,501,835 – 26,501,928 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 66.70 |
| Shannon entropy | 0.67278 |
| G+C content | 0.57506 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26501835 93 - 27905053 ------------AGUCUCCAAGUCCGAACCAAAAUCCGGAGUUCUGGGCCAAAGAGACGGAGGCGGAGACGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU ------------.((((((...((((........(((((....))))).........))))...)))))).((((((((((.........))))))))))..... ( -37.63, z-score = -2.88, R) >droSim1.chr3R 26148935 93 - 27517382 ------------AGUCUCCAAGUCCGAACCAAAAUCCGGAGUCCUGGGCCAAAGAGACGGAGGCGGAGGCGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU ------------.((((((...((((..........))))((((((...........))).))))))))).((((((((((.........))))))))))..... ( -36.80, z-score = -1.94, R) >droSec1.super_4 5328326 93 - 6179234 ------------AGUCUCCAAAUCCGAACCAAAAUCCGGAGUCCUGGACCAAAGAGACGGAGGCGGAGGCGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU ------------.((((((...((((........(((((....))))).........))))...)))))).((((((((((.........))))))))))..... ( -38.43, z-score = -3.15, R) >droYak2.chr3R 27427539 93 - 28832112 ------------AGUCUCCAAGUAUGAACCAAAGUCCGGAGUCCUGGACCAAAGAGACGGAGGCGGAGACGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU ------------.((((((..((....))....((((((....)))))).........))))))(....).((((((((((.........))))))))))..... ( -36.70, z-score = -3.38, R) >droAna3.scaffold_12911 882804 105 + 5364042 AGUCUCCAAGUCCUUGUCCGUGUCCGUGUUGGUGUCCGCGUCCGAGAUCCAAGGAGAUGGAGUCGGAGACGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU .((((((.(.(((.(.(((.((...((.((((((....)).)))).)).)).))).).))).).)))))).((((((((((.........))))))))))..... ( -43.80, z-score = -3.42, R) >dp4.chr2 30093606 89 + 30794189 -------------UCCAAGGAGAUGGAGACAGAGAAUGGACUCGGAGGC---AGAGGCGGAGUCGGAGACGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU -------------(((.......((....))(((......))))))(((---(...((((.((((....))))..)))).))))...........(((....))) ( -33.00, z-score = -3.53, R) >droPer1.super_7 959576 89 + 4445127 -------------UCCAAGGAGAUGGAGACAGAGAAUGGACUCGGAGGC---GGAGGCGGAGUCGGAGACGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU -------------(((.......((....))(((......))))))(((---(...((((.((((....))))..)))).))))...........(((....))) ( -32.30, z-score = -2.99, R) >droMoj3.scaffold_6540 3692724 100 + 34148556 --UUUGGGAGUCGGAGUCGGAGUCAGUUCCAGUGCC--AGUGCCAG-UUCCAGCUCGAGCCACCAAAGACGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU --(((((..((..((((.((((...((.(.......--.).))...-)))).))))..))..)))))....((((((((((.........))))))))))..... ( -33.10, z-score = -0.83, R) >droVir3.scaffold_13047 11295268 100 - 19223366 --GUUGGGAGUCGGAGUCGGAGUCAGUUCCAGUUCC--AGUUCUAG-UUCGAGUUUAAGGCGCCAAAGACGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU --(((.((.(((((((..((((....))))..))))--..(((...-...))).....))).))...))).((((((((((.........))))))))))..... ( -34.80, z-score = -2.29, R) >droGri2.scaffold_14624 1294876 88 - 4233967 -------UGGUCCGAUUCG----CAGUCCGAUUCCC--AGUUCGAG-UUCGAG---GCGCCACCAAAGACGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU -------((((.((.((((----.((..(((.....--...)))..-))))))---.))..))))......((((((((((.........))))))))))..... ( -28.80, z-score = -0.79, R) >consensus ____________AGUCUCCGAGUCCGAACCAGAGUCCGGAGUCCAGGGCCAAAGAGACGGAGGCGGAGACGAUGGCCGCAUGCCGAUAAUGUGCGGCCAUAAGGU .......................................................................((((((((((.........))))))))))..... (-17.40 = -17.40 + 0.00)
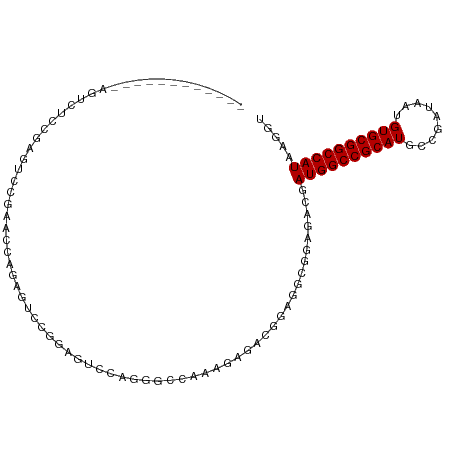
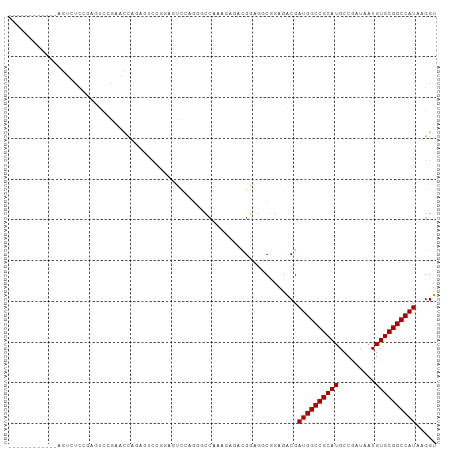
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:52 2011