
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,236,519 – 9,236,604 |
| Length | 85 |
| Max. P | 0.912472 |

| Location | 9,236,519 – 9,236,604 |
|---|---|
| Length | 85 |
| Sequences | 12 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 91.71 |
| Shannon entropy | 0.19487 |
| G+C content | 0.54379 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -22.00 |
| Energy contribution | -21.82 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9236519 85 + 23011544 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACUUGUUUGC ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).......... ( -25.20, z-score = -1.85, R) >droSim1.chr2L 9012507 85 + 22036055 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACUUGUCUUC ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).......... ( -25.20, z-score = -2.15, R) >droSec1.super_3 4695467 85 + 7220098 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACUUGCCUUC ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).......... ( -25.20, z-score = -2.17, R) >droYak2.chr2L 11903286 85 + 22324452 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACUUGCCUUC ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).......... ( -25.20, z-score = -2.17, R) >droEre2.scaffold_4929 9841877 85 + 26641161 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACCUGUCUUC ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).......... ( -25.20, z-score = -2.11, R) >droAna3.scaffold_12943 576606 85 + 5039921 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACUUGUUAAC ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).......... ( -25.20, z-score = -2.42, R) >dp4.chr4_group5 1554377 85 + 2436548 AAGGCCCCGACGAGAUUCGAACUCAUGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCGUUGACGAU ..(((.(((..((((((.........))))))(((((.....)))))(.((((.........))))).))).)))((....)).. ( -19.90, z-score = -0.81, R) >droPer1.super_5 5962452 85 + 6813705 AAGGCCCCGACGAGAUUCGAACUCAUGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCGUUGACGAU ..(((.(((..((((((.........))))))(((((.....)))))(.((((.........))))).))).)))((....)).. ( -19.90, z-score = -0.81, R) >droMoj3.scaffold_6496 21931485 83 - 26866924 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCCACUGGU-- ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........-- ( -24.30, z-score = -1.41, R) >droVir3.scaffold_10324 1107857 85 + 1288806 CAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCCUCGUGAUUU ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).......... ( -24.30, z-score = -1.56, R) >droWil1.scaffold_180698 9356226 84 - 11422946 -AGGCCCCGACGAGAUUCGAACUCAUGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCGAUGCUCUUC -.(((.(((..((((((.........))))))(((((.....)))))(.((((.........))))).))).))).......... ( -20.70, z-score = -0.96, R) >anoGam1.chr2R 51286919 85 - 62725911 UAGGCUCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAGUUGUGUGC ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).......... ( -25.70, z-score = -1.09, R) >consensus AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACUUGUCUUC ..(((.(((..(((((.((......)))))))(((((.....)))))(.((((.........))))).))).))).......... (-22.00 = -21.82 + -0.19)

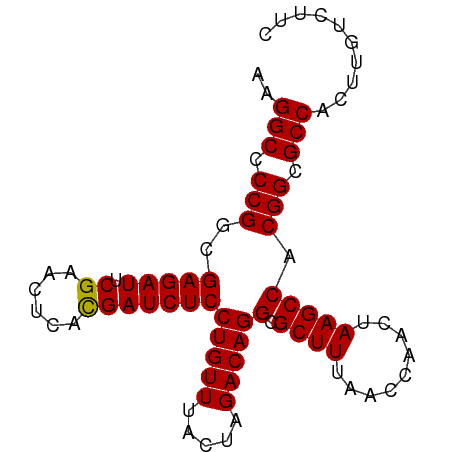
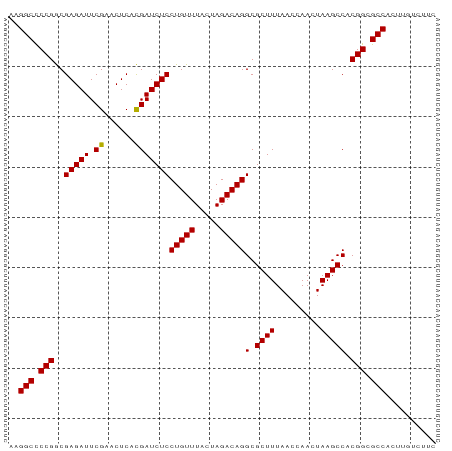
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:50 2011