| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,484,491 – 26,484,589 |
| Length | 98 |
| Max. P | 0.987121 |

| Location | 26,484,491 – 26,484,589 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 69.16 |
| Shannon entropy | 0.67566 |
| G+C content | 0.45579 |
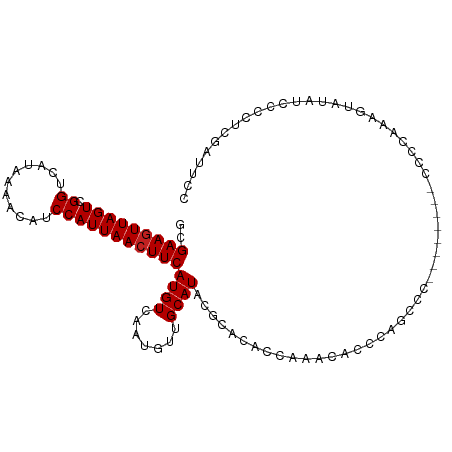
| Mean single sequence MFE | -15.20 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.09 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.987121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
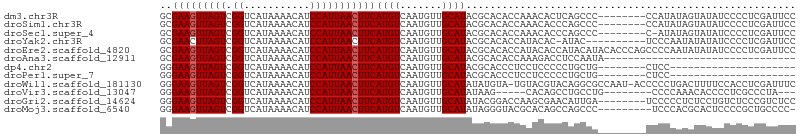
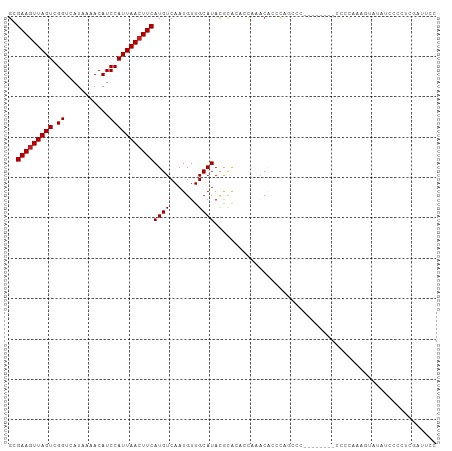
>dm3.chr3R 26484491 98 + 27905053 GCGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUACGCACACCAAACACUCAGCCC--------CCAUAUAGUAUAUCCCCUCGAUUCC (((((((((((.((...........))))))))))).(((...(((.((....)))))....)))....))..--------......................... ( -14.20, z-score = -1.80, R) >droSim1.chr3R 26130293 98 + 27517382 GCGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUACGCACACCAAACACCCAGCCC--------CCAUAUAGUAUAUCCCCUCGAUUCC (((((((((((.((...........))))))))))).(((...(((.((....)))))....)))....))..--------......................... ( -14.20, z-score = -1.95, R) >droSec1.super_4 5311015 97 + 6179234 GCGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUACGCACACCAAACACCCAGCCC--------C-AUAUAGUAUAUCCCCUCGAUUCC (((((((((((.((...........))))))))))).(((...(((.((....)))))....)))....))..--------.-....................... ( -14.20, z-score = -1.91, R) >droYak2.chr3R 27408765 95 + 28832112 GCGAACUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUACGCACACCAUACAC-AUAC----------UCCCAAUAUAUAUCCCCUCGAUUCC ..(((.(((((.((...........))))))).)))((((..(((.(((....)))...)))..))-))..----------......................... ( -8.00, z-score = 0.17, R) >droEre2.scaffold_4820 8957470 106 - 10470090 GCGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUACGCACACCAUACACCAUACAUACACCCAGCCCCAAUAUAUAUCCCCUCGAUUCC (((((((((((.((...........))))))))))).(((...(((.((....)))))....)))...............))........................ ( -14.20, z-score = -1.65, R) >droAna3.scaffold_12911 865516 74 - 5364042 GCGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUACGCACACCAAAGACCUCCAAUA-------------------------------- ..(((((((((.((...........)))))))))))..(((..((.(((....)))...))..)))........-------------------------------- ( -13.80, z-score = -2.30, R) >dp4.chr2 30073498 77 - 30794189 GGGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUACGCACCCUCCUCCCCCUGCUG--------CUCC--------------------- (((((((((((.((...........)))))))))))..........(((....))).))..............--------....--------------------- ( -13.10, z-score = -0.56, R) >droPer1.super_7 939333 77 - 4445127 GGGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUACGCACCCUCCUCCCCCUGCUG--------CUCC--------------------- (((((((((((.((...........)))))))))))..........(((....))).))..............--------....--------------------- ( -13.10, z-score = -0.56, R) >droWil1.scaffold_181130 10001872 104 + 16660200 GGGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUAUAUGUA-UGUACGUACAGGCGCCAAU-ACCCCCUGACUUUUCCACCUCGAUUUC (..(((((((..(((...........((((.((.....)).))))..((.....((((-(....))))).))......-)))..)))))))..)............ ( -21.40, z-score = -0.84, R) >droVir3.scaffold_13047 11273677 90 + 19223366 GGGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUAUAAG-----CACAGCCUGCCUG--------CCCCAAACACCCCUCGCCCUA--- (((((((((((.((...........))))))))))).......(((.((......)-----)))).........--------..)).................--- ( -14.70, z-score = 0.13, R) >droGri2.scaffold_14624 1265475 98 + 4233967 GGGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUAUACGGACCAAGCGAACAUUGA--------UCCCCCUCUCCUGUCUCCCGUCUCC (((((((((((.((...........)))))))))))..(((((((((((.............)).))))))))--------)................))...... ( -21.32, z-score = -1.45, R) >droMoj3.scaffold_6540 3670617 96 - 34148556 GGGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUAUAGGGUACGCACAGCCAGCCC---------UCCCACGCACUCCCCGCUGCCCC- (((((((((((.((...........)))))))))))..........(((....(((((..((...))..))))---------).....)))....))........- ( -20.20, z-score = 0.06, R) >consensus GCGAAGUUAGUCGGUCAUAAAACAUCCAUUAACUUCAUGUCAAUGUUGCAUACGCACACCAAACACCCAGCCC________CCCCAAAGUAUAUCCCCUCGAUUCC ..(((((((((.((...........)))))))))))((((.......))))....................................................... (-10.01 = -10.09 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:48 2011