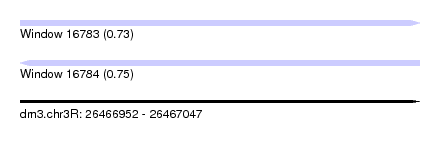
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,466,952 – 26,467,047 |
| Length | 95 |
| Max. P | 0.747465 |

| Location | 26,466,952 – 26,467,047 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.15 |
| Shannon entropy | 0.25597 |
| G+C content | 0.34668 |
| Mean single sequence MFE | -18.76 |
| Consensus MFE | -16.11 |
| Energy contribution | -16.11 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734870 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
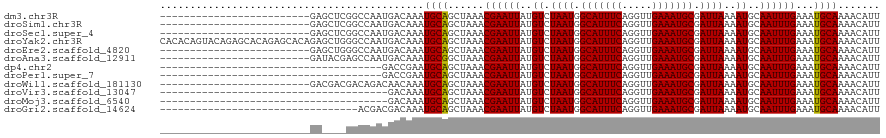
>dm3.chr3R 26466952 95 + 27905053 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCAUUGGCCGAGCUC------------------------- .(((((.((((........))))...((((.(((((((.....))))))).))))))))).....(((..(((((....))....)))..)))..------------------------- ( -20.70, z-score = -1.32, R) >droSim1.chr3R 26112893 95 + 27517382 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCAUUGGCCGAGCUC------------------------- .(((((.((((........))))...((((.(((((((.....))))))).))))))))).....(((..(((((....))....)))..)))..------------------------- ( -20.70, z-score = -1.32, R) >droSec1.super_4 5293723 95 + 6179234 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCAUUGGCCGAGCUC------------------------- .(((((.((((........))))...((((.(((((((.....))))))).))))))))).....(((..(((((....))....)))..)))..------------------------- ( -20.70, z-score = -1.32, R) >droYak2.chr3R 27389139 120 + 28832112 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCAUUGGCCCAGCUCUGUGCUCUGUGCUCUGUACUGUGUG .(((((.((((........))))...((((.(((((((.....))))))).))))))))).....((.(((..((((..(.(((.(((...)))..))))...)))).))).))...... ( -24.60, z-score = -0.25, R) >droEre2.scaffold_4820 8939844 95 - 10470090 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCAUUGGCCCAGCUC------------------------- .(((((.((((........))))...((((.(((((((.....))))))).))))))))).........(((((.....((.....)).))))).------------------------- ( -19.70, z-score = -1.31, R) >droAna3.scaffold_12911 849303 95 - 5364042 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCCGCAUUUGUCAUUGGCUCGUAUC------------------------- .(((((.((((........))))...((((.(((((((.....))))))).))))))))).........(((((..........)))))......------------------------- ( -19.30, z-score = -1.60, R) >dp4.chr2 30055138 83 - 30794189 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUCGGUC------------------------------------- .(((((.((((........))))...((((.(((((((.....))))))).)))))))))..........((((....)))).------------------------------------- ( -16.40, z-score = -1.56, R) >droPer1.super_7 920038 83 - 4445127 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUCGGUC------------------------------------- .(((((.((((........))))...((((.(((((((.....))))))).)))))))))..........((((....)))).------------------------------------- ( -16.40, z-score = -1.56, R) >droWil1.scaffold_181130 9983217 95 + 16660200 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUUGUCUGUCGUCGUC------------------------- .(((....(((...(((((.(((........(((((((.....)))))))..((((((.......)))))).)))))))).)))....)))....------------------------- ( -17.40, z-score = -0.95, R) >droVir3.scaffold_13047 11246374 82 + 19223366 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUC-------------------------------------- (((((...((....(.(((((...((((((.(((((((.....))))))).))))))..))))).)....)).)))))....-------------------------------------- ( -16.20, z-score = -1.67, R) >droMoj3.scaffold_6540 3649623 82 - 34148556 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUC-------------------------------------- (((((...((....(.(((((...((((((.(((((((.....))))))).))))))..))))).)....)).)))))....-------------------------------------- ( -16.20, z-score = -1.67, R) >droGri2.scaffold_14624 1226668 87 + 4233967 AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCGUCGU--------------------------------- .......((((........))))...((((.(((((((.....))))))).))))(((((((...((......))..)))).)))..--------------------------------- ( -16.80, z-score = -1.33, R) >consensus AAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCAUUGGCC____UC_________________________ (((((..((((........)))).((((((.(((((((.....))))))).))))))................))))).......................................... (-16.11 = -16.11 + -0.00)


| Location | 26,466,952 – 26,467,047 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.15 |
| Shannon entropy | 0.25597 |
| G+C content | 0.34668 |
| Mean single sequence MFE | -18.91 |
| Consensus MFE | -18.02 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747465 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
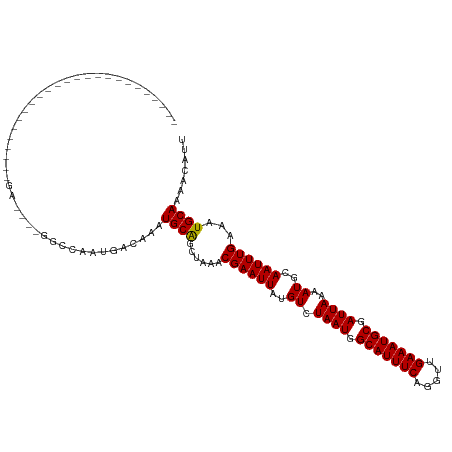
>dm3.chr3R 26466952 95 - 27905053 -------------------------GAGCUCGGCCAAUGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU -------------------------..((...((((..(((((((.(........).))).))))...))))(((((((((((................)))))))))))))........ ( -18.39, z-score = -0.04, R) >droSim1.chr3R 26112893 95 - 27517382 -------------------------GAGCUCGGCCAAUGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU -------------------------..((...((((..(((((((.(........).))).))))...))))(((((((((((................)))))))))))))........ ( -18.39, z-score = -0.04, R) >droSec1.super_4 5293723 95 - 6179234 -------------------------GAGCUCGGCCAAUGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU -------------------------..((...((((..(((((((.(........).))).))))...))))(((((((((((................)))))))))))))........ ( -18.39, z-score = -0.04, R) >droYak2.chr3R 27389139 120 - 28832112 CACACAGUACAGAGCACAGAGCACAGAGCUGGGCCAAUGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU ......((.((..((.(((..(...)..))).))...))))...((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))....... ( -23.70, z-score = 0.17, R) >droEre2.scaffold_4820 8939844 95 + 10470090 -------------------------GAGCUGGGCCAAUGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU -------------------------.(((((.(...........).))))).........((((.((((.(((((((.....))))))).))))...((((........))))..)))). ( -19.90, z-score = -0.51, R) >droAna3.scaffold_12911 849303 95 + 5364042 -------------------------GAUACGAGCCAAUGACAAAUGCGGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU -------------------------.......((((..(((((((.((......)).))).))))...))))(((((((((((................))))))))))).......... ( -20.19, z-score = -1.28, R) >dp4.chr2 30055138 83 + 30794189 -------------------------------------GACCGAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU -------------------------------------.......((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))....... ( -18.00, z-score = -1.62, R) >droPer1.super_7 920038 83 + 4445127 -------------------------------------GACCGAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU -------------------------------------.......((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))....... ( -18.00, z-score = -1.62, R) >droWil1.scaffold_181130 9983217 95 - 16660200 -------------------------GACGACGACAGACAACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU -------------------------...................((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))....... ( -18.00, z-score = -1.20, R) >droVir3.scaffold_13047 11246374 82 - 19223366 --------------------------------------GACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU --------------------------------------......((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))....... ( -18.00, z-score = -1.77, R) >droMoj3.scaffold_6540 3649623 82 + 34148556 --------------------------------------GACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU --------------------------------------......((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))....... ( -18.00, z-score = -1.77, R) >droGri2.scaffold_14624 1226668 87 - 4233967 ---------------------------------ACGACGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU ---------------------------------...........((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))....... ( -18.00, z-score = -1.41, R) >consensus _________________________GA____GGCCAAUGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUU ............................................((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))....... (-18.02 = -17.94 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:46 2011