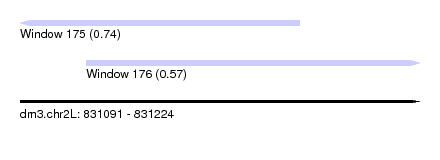
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 831,091 – 831,224 |
| Length | 133 |
| Max. P | 0.735209 |

| Location | 831,091 – 831,184 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.96 |
| Shannon entropy | 0.09666 |
| G+C content | 0.34165 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 831091 93 - 23011544 AGCGCGACCUAUAUAUUUGAAAAUAGUUGUUAUUAGCCAAUAAUGACAACCUCUCAUAGAUGAUGUGAAUG--UUUAUUUUGAGAACACGUCCAG .(.(((...................((((((((((.....)))))))))).(((((.((((((........--.)))))))))))...))).).. ( -18.70, z-score = -2.12, R) >droSim1.chr2L 828962 94 - 22036055 AGCGCGACCUAUAUAGUUGAAAAUAAUUGUUAUUAGCCAAUAAUGACAACCUCUUA-AGAUGAUGUGAAUGGACUUAUUUUGAGAACACGUCGAU ....((((..................(((((((((.....)))))))))..(((((-((((((.((......)))))))))))))....)))).. ( -20.60, z-score = -2.83, R) >droSec1.super_14 801495 94 - 2068291 AGGGCGACCUAUAUAGUUGAAAAUAAUUGUUAUUAGCCAAUAAUGACAACCUCUUA-AGAUGAUGUGAAUGGACUUAUUUUGAGAACACGUCCAU .(((((....................(((((((((.....)))))))))..(((((-((((((.((......)))))))))))))...))))).. ( -24.40, z-score = -3.65, R) >consensus AGCGCGACCUAUAUAGUUGAAAAUAAUUGUUAUUAGCCAAUAAUGACAACCUCUUA_AGAUGAUGUGAAUGGACUUAUUUUGAGAACACGUCCAU .....(((..................(((((((((.....)))))))))..(((((.((((((.((......)))))))))))))....)))... (-13.84 = -13.73 + -0.11)

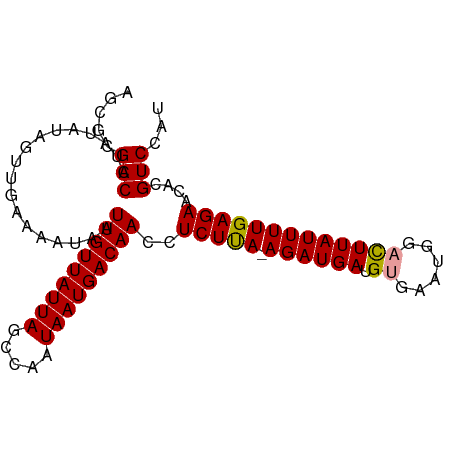
| Location | 831,113 – 831,224 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 60.88 |
| Shannon entropy | 0.75690 |
| G+C content | 0.45999 |
| Mean single sequence MFE | -21.28 |
| Consensus MFE | -9.07 |
| Energy contribution | -8.80 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 831113 111 + 23011544 -CAUUCACAUCAUCUAUGAGAGGUUGUCAUUAUUGGCUAAUAACAACUAUUUUC----AAAUAUAUAGGUCGCGCUGUACAGCUUCACGUCAAACAACGACCAGGAGCUCUCGUUC -..............((((((((((((.((((.....)))).))))).......----.........(((((.(((....))).....(.....)..))))).....))))))).. ( -22.80, z-score = -1.57, R) >droSim1.chr2L 828985 111 + 22036055 CCAUUCACAUCAUCU-UAAGAGGUUGUCAUUAUUGGCUAAUAACAAUUAUUUUC----AACUAUAUAGGUCGCGCUGUACAGCUUCACGUCGAACAACGACCAGGAGCUCUCGUUC ...............-...(((((((.....((((........))))......)----)))..(((((......))))).((((((..((((.....))))..))))))))).... ( -20.50, z-score = -0.80, R) >droSec1.super_14 801518 111 + 2068291 CCAUUCACAUCAUCU-UAAGAGGUUGUCAUUAUUGGCUAAUAACAAUUAUUUUC----AACUAUAUAGGUCGCCCUGUACAGCUUCACGUCGAACAACGACCAGGAGCUCUCGUUC ...............-...(((((((.....((((........))))......)----)))..((((((....)))))).((((((..((((.....))))..))))))))).... ( -23.20, z-score = -1.95, R) >droEre2.scaffold_4929 887877 100 + 26641161 CCAUUCUCAUCA--------AGAUUAU----AUAUGGUAAUAACAAUUCUUUUC----ACCAAUAUAGGUCGCCCUUUACAGCUUCACGUCGAACAACGACCAGGAGCUCUCGUUC ............--------.((((((----((.((((................----))))))))).))).........((((((..((((.....))))..))))))....... ( -15.99, z-score = -1.26, R) >droAna3.scaffold_12916 2721728 99 + 16180835 -----CAGAUCCCUC----AAGACUAA----ACUGGAUAGUAACUCCCCAUCCC----CCAUCCACAGGUUGCUCUCUACAGCUUCACGUCAAACAACGACCAGGAGCUCUCGUUC -----..........----..(((...----..((((.(((((((.........----.........))))))).)))).((((((..(((.......)))..))))))...))). ( -16.47, z-score = -0.13, R) >dp4.chr4_group3 5623728 94 + 11692001 ----------------UCUAAUGUGGUGGCCUCUAAUGGACGGACGUUCUCU------UCUCUUGCAGGUGGCCCUCUACAGCUUCACGUCGAACAAUGACCAGGAGCUGUCGUUU ----------------........(((.((((.(((.(((.(((.....)))------))).))).)))).)))....((((((((..((((.....))))..))))))))..... ( -26.40, z-score = -0.15, R) >droPer1.super_1 7115129 94 + 10282868 ----------------UCUAAUGUGGUGGCCUCUAAUGGACGGACGUUCUCU------UCUCUUGCAGGUGGCCCUCUACAGCUUCACGUCGAACAAUGACCAGGAGCUGUCGUUU ----------------........(((.((((.(((.(((.(((.....)))------))).))).)))).)))....((((((((..((((.....))))..))))))))..... ( -26.40, z-score = -0.15, R) >droVir3.scaffold_12963 19365903 93 + 20206255 --------------------AAAUGUAUACCUUAUCUUAAGCUAUCCUUUCAACG---UCGCUUGCAGGUGGCGCUCUAUAGCUUCACGUCGAACAAUGACCAGGAGCUGUCGUUU --------------------..................((((...........((---(((((....)))))))....((((((((..((((.....))))..)))))))).)))) ( -19.60, z-score = 0.04, R) >droMoj3.scaffold_6500 8507548 91 - 32352404 -----------------GCCAAGCUAU--CCUCAUCUCAAUCUGUCUUUAUC------UUGCUUGCAGGUGGCGCUCUAUAGCUUCACAUCGAACAAUGAUCAGGAGCUGUCGUUU -----------------.....((((.--(((((...(((..((....))..------)))..)).))))))).....((((((((..((((.....))))..))))))))..... ( -17.00, z-score = 0.84, R) >droGri2.scaffold_15252 7239608 99 + 17193109 -----------------ACCACGCGAUGGGCUUCUCUUAAUCACUGCUUAACACACUCUUCCUUGCAGGUGGCGCUGUAUAGCUUCACAUCGAACAAUGAUCAGGAGCUGUCGUUU -----------------.....(((((((.(((((....((((((((.................))))((((.(((....))).)))).........)))).)))))))))))).. ( -24.43, z-score = -0.61, R) >consensus ___________________AAGGUUGU_ACCAUUGGUUAAUAACAAUUCUUUUC____UCCCUUACAGGUGGCGCUCUACAGCUUCACGUCGAACAACGACCAGGAGCUCUCGUUC ................................................................................((((((..((((.....))))..))))))....... ( -9.07 = -8.80 + -0.27)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:05 2011