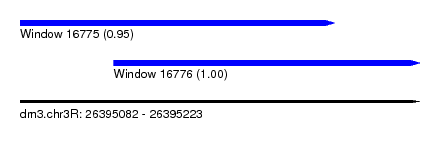
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,395,082 – 26,395,223 |
| Length | 141 |
| Max. P | 0.999249 |

| Location | 26,395,082 – 26,395,193 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.92 |
| Shannon entropy | 0.45575 |
| G+C content | 0.49350 |
| Mean single sequence MFE | -36.57 |
| Consensus MFE | -21.47 |
| Energy contribution | -23.43 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
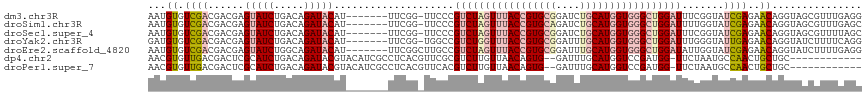
>dm3.chr3R 26395082 111 + 27905053 AAUGUGUCGACGACGAGUAUCUGACAGAUACAU-------UUCGG-UUCCCGUCUAGUUUACCGUGCGGAUCUGCAUGGUGGGCUGGAUUUCGGUAUCGAGAACAGGUAGCGUUUGAGG ......((((((.(..(((((.....))))).(-------(((((-(.((.(((((((((((((((((....)))))))))))))))))...)).))))))).......))).)))).. ( -41.50, z-score = -2.67, R) >droSim1.chr3R 26040472 111 + 27517382 AAUGUGUCGACGACGAGUAUCUGACAGAUACAU-------UUCGG-UUCCCGUCUAGUUUACCGUGCAGAUCUGCAUGGUGGGCUGGAUUUUGGUAUCGAGAACAGGUAGCGUUUGAGC ......((((((.(..(((((.....))))).(-------(((((-(.((.(((((((((((((((((....)))))))))))))))))...)).))))))).......))).)))).. ( -42.30, z-score = -3.48, R) >droSec1.super_4 5221809 111 + 6179234 AAUGUGUCGACGACGAGUAUCUGACAGAUACAU-------UUCGG-UUCCCGUCUAGUUUACCGUGCGGAUCUGCAUGGUGGGCUGGAUUUCGGUAUCGAGAACAGGUAGCGUUUUAGC ...((...((((.(..(((((.....))))).(-------(((((-(.((.(((((((((((((((((....)))))))))))))))))...)).))))))).......)))))...)) ( -41.20, z-score = -2.91, R) >droYak2.chr3R 27314636 111 + 28832112 GAUGUGUCGACGACGAGUAUCUGACAGAUACAU-------UUCGG-UGGCCGUCUGGUUUACCGUGCGGAUUUGCAUGGUGGGCUGGAUUUGGGUAUUGAGAACAGGUAUCUUUUCAGG ....((((...)))).....((((.((((((.(-------(((((-((.(((((..((((((((((((....))))))))))))..)))..)).))))))))....))))))..)))). ( -43.00, z-score = -3.81, R) >droEre2.scaffold_4820 8860086 112 - 10470090 AAUGUGUCGACGACGAGUAUCUGGCAGAUACAU-------UUCGGCUUGCCGUCUAGUUUACCGUGCGGAUUUGCAUGGUGGGCUGGAUAUUGGUAUCGAGAACAGGUAUCUUUUGAGG ..........(((.(((((((((...(((((..-------...((....))(((((((((((((((((....)))))))))))))))))....))))).....)))))).))))))... ( -40.00, z-score = -2.89, R) >dp4.chr2 29657408 104 - 30794189 AACGUGUUGACGACUCGCAUCUGACAGAUACGUACAUCGCCUCACGUUCGCGUCUUGUUAACAGUG--GAUUUGCAUGGUCCGAUGG-UUCUAAUGCCAACUGCUGC------------ .((.(((((((((..((((((.....)))((((..........))))..)))..)))))))))))(--((((.....)))))..(((-(......))))........------------ ( -24.70, z-score = 1.04, R) >droPer1.super_7 515439 104 - 4445127 AACGUGUUGACGACUCGCAUCUGACAGAUACGUACAUCGCCUCACGUUCACGUCUUGUUAACAGUG--GAUUUGCAUGGUCCGAUGG-UUCUAAUGCCAACUGCUGC------------ ((((((..(.(((..((.(((.....))).))....))))..)))))).............(((((--((((.....)))))..(((-(......))))...)))).------------ ( -23.30, z-score = 1.19, R) >consensus AAUGUGUCGACGACGAGUAUCUGACAGAUACAU_______UUCGG_UUCCCGUCUAGUUUACCGUGCGGAUUUGCAUGGUGGGCUGGAUUUUGGUAUCGAGAACAGGUAGCGUUUGAGG ...((.((((......(((((.....)))))....................(((((((((((((((((....))))))))))))))))).......))))..))............... (-21.47 = -23.43 + 1.96)
| Location | 26,395,115 – 26,395,223 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.82 |
| Shannon entropy | 0.59172 |
| G+C content | 0.48382 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -16.36 |
| Energy contribution | -19.81 |
| Covariance contribution | 3.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
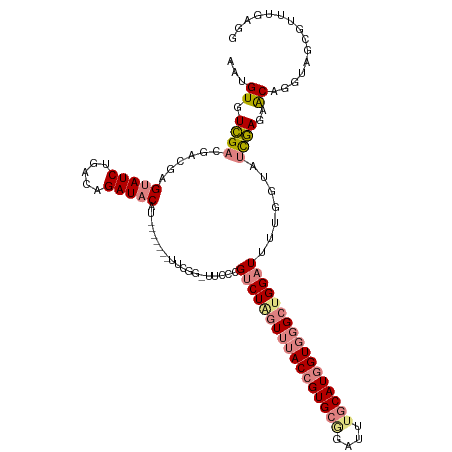
>dm3.chr3R 26395115 108 + 27905053 UUCGG-UUCCCGUCUAGUUUACCGUGCGGAUCUGCAUGGUGGGCUGGAUUUCGGUAUCGAGAACAGGUAGCGUUUGAGGUACUCUAUUUUAACAGGAAGCAUUACCCCA- ...(.-((((.(((((((((((((((((....)))))))))))))))))...((((((..((((.......))))..))))))...........)))).).........- ( -38.50, z-score = -2.48, R) >droSim1.chr3R 26040505 108 + 27517382 UUCGG-UUCCCGUCUAGUUUACCGUGCAGAUCUGCAUGGUGGGCUGGAUUUUGGUAUCGAGAACAGGUAGCGUUUGAGCUACUCUACUUGAACGGGGAACAUUAUACCA- ....(-((((((((((((((((((((((....)))))))))))))))))....((.(((((....((((((......))))))...))))))).)))))).........- ( -48.50, z-score = -5.72, R) >droSec1.super_4 5221842 108 + 6179234 UUCGG-UUCCCGUCUAGUUUACCGUGCGGAUCUGCAUGGUGGGCUGGAUUUCGGUAUCGAGAACAGGUAGCGUUUUAGCUACUCUACUUGAACAGGGAACAUUAUACCA- ....(-((((((((((((((((((((((....)))))))))))))))))....((.(((((....((((((......))))))...))))))).)))))).........- ( -47.90, z-score = -5.70, R) >droYak2.chr3R 27314669 109 + 28832112 UUCGG-UGGCCGUCUGGUUUACCGUGCGGAUUUGCAUGGUGGGCUGGAUUUGGGUAUUGAGAACAGGUAUCUUUUCAGGUGCUCUAUUUGAACGGGUAAUUUGAUAGCUG (((((-((.(((((..((((((((((((....))))))))))))..)))..)).)))))))..(((((((((.((((((((...)))))))).)))).)))))....... ( -40.40, z-score = -3.93, R) >droEre2.scaffold_4820 8860119 110 - 10470090 UUCGGCUUGCCGUCUAGUUUACCGUGCGGAUUUGCAUGGUGGGCUGGAUAUUGGUAUCGAGAACAGGUAUCUUUUGAGGUACUCUAUUUUAACGGGAAACAUUAUACCAG ...((....))(((((((((((((((((....)))))))))))))))))..((((((..((((.((((((((....)))))).)).))))....(....)...)))))). ( -39.60, z-score = -4.03, R) >dp4.chr2 29657448 95 - 30794189 CUCACGUUCGCGUCUUGUUAACAGUG--GAUUUGCAUGGUCCGAUGG-UUCUAAUGCCAACUGCUGCUGCCCUC-------CUUUGGGGGAACGAACGAUUUCGA----- ....((((((......((...(((((--((((.....)))))..(((-(......))))))))..))..(((((-------....)))))..)))))).......----- ( -27.00, z-score = -0.51, R) >droPer1.super_7 515479 95 - 4445127 CUCACGUUCACGUCUUGUUAACAGUG--GAUUUGCAUGGUCCGAUGG-UUCUAAUGCCAACUGCUGCUGCCCUC-------CUUUGGGAGAACGAACGAUUUCGA----- ....(((((.......((...(((((--((((.....)))))..(((-(......))))))))..))....(((-------(....))))...))))).......----- ( -24.40, z-score = -0.35, R) >consensus UUCGG_UUCCCGUCUAGUUUACCGUGCGGAUUUGCAUGGUGGGCUGGAUUUUGGUAUCGAGAACAGGUAGCGUUUGAGGUACUCUAUUUGAACGGGAAACAUUAUACCA_ ...........(((((((((((((((((....)))))))))))))))))................((((((......))))))........................... (-16.36 = -19.81 + 3.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:38 2011