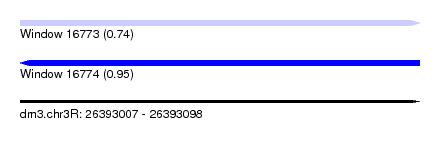
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,393,007 – 26,393,098 |
| Length | 91 |
| Max. P | 0.945389 |

| Location | 26,393,007 – 26,393,098 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 81.86 |
| Shannon entropy | 0.31674 |
| G+C content | 0.53180 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -21.77 |
| Energy contribution | -22.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.736316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26393007 91 + 27905053 -GAUUCUGGUUGCAACGAUGCGGCAGAGACCGAGGCUUAUACCCUGUACUUAGGCAGAACAAGGGUAUGCUCGGUGAUGCUGCUGCCCAA-UA-- -.....(((..(((.....((((((...((((((...((((((((((.((.....)).)).))))))))))))))..)))))))))))).-..-- ( -32.80, z-score = -1.95, R) >droSim1.chr3R 26038402 94 + 27517382 -GAUUCUGGUUGCAACGAUGCGGCAGAGACCGAGGCCUAUACCCUGUACUUAGACAGAAGAAGGGUAUGCUCGGUGAUGCUGCUGCCCAAAUAAG -.....(((..(((.....((((((...((((((...((((((((...(((......))).))))))))))))))..))))))))))))...... ( -32.70, z-score = -2.64, R) >droSec1.super_4 5219771 91 + 6179234 -GAUUCUGGUUGCAACGAUGCGGCAGAGACCGAGGCCUAUACCCUGUACUUAGACGGAACAAGGGAAUGCUCGGUGAUGCUGCUGCCUAAAA--- -..................(((((((..((((((...(((.((((((.((.....)).)).)))).)))))))))....)))))))......--- ( -27.10, z-score = -1.13, R) >droYak2.chr3R 27312587 88 + 28832112 GAUUUCUGGUUGCAACGAUGCGGCAGAGACGGAGGUCUAUACCCUGUGCUUAGAGCGAACUAGGGUAUGCUGGGUGAUGCAAGUAAGA------- ......((.(((((.(.((.((((..((((....))))(((((((((((.....)))...)))))))))))).))).))))).))...------- ( -28.60, z-score = -1.63, R) >droEre2.scaffold_4820 8857987 84 - 10470090 GAUUUCUGGUUGCAACGAUGCGGCAGAGACCGAGGUCUAUACCCUGCACUUUGAGGGAACGAGGGUA-GCUCGGCGAUGCUGCUG---------- ...................((((((..(.(((((.....(((((((..((.....))..).))))))-.))))))..))))))..---------- ( -26.40, z-score = -0.37, R) >consensus _GAUUCUGGUUGCAACGAUGCGGCAGAGACCGAGGCCUAUACCCUGUACUUAGACAGAACAAGGGUAUGCUCGGUGAUGCUGCUGCCCAA_____ ...(((((.(((((....))))))))))((((((...((((((((................)))))))))))))).................... (-21.77 = -22.77 + 1.00)

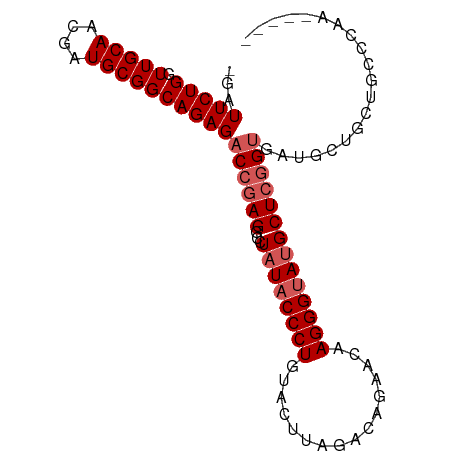
| Location | 26,393,007 – 26,393,098 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 81.86 |
| Shannon entropy | 0.31674 |
| G+C content | 0.53180 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -18.82 |
| Energy contribution | -20.54 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
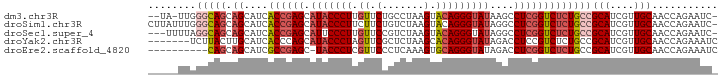
>dm3.chr3R 26393007 91 - 27905053 --UA-UUGGGCAGCAGCAUCACCGAGCAUACCCUUGUUCUGCCUAAGUACAGGGUAUAAGCCUCGGUCUCUGCCGCAUCGUUGCAACCAGAAUC- --..-((((((((.((....((((((((((((((.((.((.....)).)))))))))..).)))))))))))))(((....)))...)))....- ( -31.80, z-score = -2.79, R) >droSim1.chr3R 26038402 94 - 27517382 CUUAUUUGGGCAGCAGCAUCACCGAGCAUACCCUUCUUCUGUCUAAGUACAGGGUAUAGGCCUCGGUCUCUGCCGCAUCGUUGCAACCAGAAUC- ....(((((((((.((....((((((((((((((....((.....))...)))))))..).)))))))))))))(((....)))...))))...- ( -31.00, z-score = -2.69, R) >droSec1.super_4 5219771 91 - 6179234 ---UUUUAGGCAGCAGCAUCACCGAGCAUUCCCUUGUUCCGUCUAAGUACAGGGUAUAGGCCUCGGUCUCUGCCGCAUCGUUGCAACCAGAAUC- ---.....(((((.((....(((((((....((((((..(......).)))))).....).)))))))))))))(((....)))..........- ( -27.20, z-score = -2.15, R) >droYak2.chr3R 27312587 88 - 28832112 -------UCUUACUUGCAUCACCCAGCAUACCCUAGUUCGCUCUAAGCACAGGGUAUAGACCUCCGUCUCUGCCGCAUCGUUGCAACCAGAAAUC -------(((...(((((..((...(((((((((.((..((.....)))))))))))((((....)))).....))...)))))))..))).... ( -19.10, z-score = -1.68, R) >droEre2.scaffold_4820 8857987 84 + 10470090 ----------CAGCAGCAUCGCCGAGC-UACCCUCGUUCCCUCAAAGUGCAGGGUAUAGACCUCGGUCUCUGCCGCAUCGUUGCAACCAGAAAUC ----------..(((((.....((((.-....))))..........((((..((((.((((....)))).)))))))).)))))........... ( -23.90, z-score = -1.62, R) >consensus _____UUGGGCAGCAGCAUCACCGAGCAUACCCUUGUUCCGUCUAAGUACAGGGUAUAGGCCUCGGUCUCUGCCGCAUCGUUGCAACCAGAAUC_ ........(((((.((....((((((.(((((((.((.(.......).)))))))))....)))))))))))))(((....)))........... (-18.82 = -20.54 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:37 2011