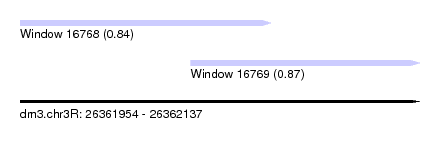
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,361,954 – 26,362,137 |
| Length | 183 |
| Max. P | 0.868333 |

| Location | 26,361,954 – 26,362,069 |
|---|---|
| Length | 115 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.41 |
| Shannon entropy | 0.48460 |
| G+C content | 0.47595 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
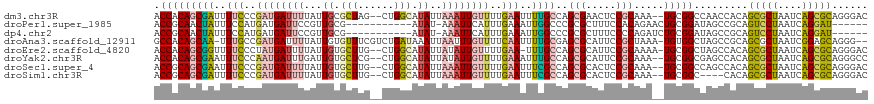
>dm3.chr3R 26361954 115 + 27905053 ACCACAGCGAUUUUCCCGAUGAUUUUAUUGCGCUAG--CUGGCAUAUUAAAUUGUUUUGAAUUUUGCCAGCGAACUCCGCAAA--UGCGGCCAACCACAGCGCUAAUCAGCGCAGGGAC .............((((..(((((.....(((((.(--((((((..(((((....)))))....))))))).....((((...--.))))........))))).))))).....)))). ( -37.40, z-score = -2.93, R) >droPer1.super_1985 5003 101 + 6095 ACCGCAACUAUUUCCAUGAUGAUUCCGUUGCG-----------AUAU-AAAUUCAUUUGAAAUUGGCCCGCGCUUUCCACAGAACUGCGGAUAGCCGCAGUCCUAAUCAGGAU------ ............(((.((((.........(((-----------...(-((.(((....))).)))...))).........((.(((((((....))))))).)).))))))).------ ( -24.10, z-score = -1.02, R) >dp4.chr2 29620988 101 - 30794189 ACCGCAACUAUUUCCAUGAUGAUUCCGUUGCG-----------AUAU-AAAUUCAUUUGAAAUUGGCCCGCGCUUUCCCCAGAUCUGCGGAUAGCCGCAGUCCUAAUCAGGAU------ ..((((((....((......))....))))))-----------....-......(((((.....(((....))).....)))))((((((....))))))((((....)))).------ ( -23.70, z-score = -0.77, R) >droAna3.scaffold_12911 741243 115 - 5364042 GCCACAGCAA-UUUGCCGAUGAUUUUAUUGUGUUUCGUCUGAUAAAUUAAUUUGUUUUCAAUUUUGCGAGCGCAUUCCGCUAAA-UGUGGCUAGCCGCAGCGCUAAUCGGAGCAGGG-- .((...((((-(((((.(((((..(......)..))))).).))))))................((((.((.....((((....-.))))...))))))))(((......))).)).-- ( -26.10, z-score = 1.28, R) >droEre2.scaffold_4820 8822556 115 - 10470090 ACCACAGCGGUUUUCCCUAUGAUUUUAUUGUGCUUG--CUGGCAUAUUAUAUUGUUUUGAA-UUUGCCAGCGCAUUCCGCAAAA-UGCGGCUAGCCACAGCGCUAAUCAGCGCAGGGAC (((.....)))..(((((..........((((...(--((((((..(((........))).-..)))))))((...((((....-.))))...))))))(((((....)))))))))). ( -39.80, z-score = -2.49, R) >droYak2.chr3R 27279657 115 + 28832112 ACCACAGCGAAUUUCCCAAUGAUUUGAUUGUGCUCG--CUGGCAUAUUAUAUUGUUUUGAAAUUUGCCAGCGCAUUCCGCAAA--UGCGGCGAGCCACAGCGCUAAUCAGCGCAGGGCC .((((((((((((.......)))))).))))(((((--((((((.(((.((......)).))).))))))).....((((...--.)))).))))....(((((....))))).))... ( -40.70, z-score = -2.30, R) >droSec1.super_4 5181300 115 + 6179234 ACCGCAGCGAAUUUCCCGAUGAUUUUAUUGUGCUUG--CUGGCAUAUUAAAUUGUUUUGAAUUUCGCCAGCGCACUCCGCAAA--UGCGGCCAGCCACAGCGCUAAUCAGCGCAGGGAC .((((((((((.(((..((..((((...((((((..--..))))))..))))..))..))).)))))..(((.....)))...--)))))....((...(((((....)))))..)).. ( -40.00, z-score = -2.20, R) >droSim1.chr3R 26007698 111 + 27517382 ACCGCAGCGAUUUUCCCGAUGAUUUUAUUGUGCUUG--CUGGCAUAUUAAAUUGUUUUGAAUUUCGCCAGCGCACUCCGCAAA--UGCGGC----CACAGCGCUAAUCAGCGCAGGGAC .(((((((((..(((..((..((((...((((((..--..))))))..))))..))..)))..))))..(((.....)))...--)))))(----(...(((((....)))))..)).. ( -39.70, z-score = -2.70, R) >consensus ACCACAGCGAUUUUCCCGAUGAUUUUAUUGUGCUUG__CUGGCAUAUUAAAUUGUUUUGAAUUUUGCCAGCGCAUUCCGCAAA__UGCGGCUAGCCACAGCGCUAAUCAGCGCAGGGAC .(((((((((..(((..((((((((.((...(((......)))..)).))))))))..)))..))))..(((.....))).....))))).........(((((....)))))...... (-17.98 = -18.32 + 0.35)

| Location | 26,362,032 – 26,362,137 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Shannon entropy | 0.39096 |
| G+C content | 0.64988 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -20.91 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26362032 105 + 27905053 AAAUGCGGCCAACCACAGCGCUAAUCAGCGCAGGGACAGCUGUCUCUGCACCACCCACUAUCCCGUCGCCUGGACCACCGACCGCACCGCCCACCGCACCACCAG---------- ...(((((.........(((((....))))).(((...((.(((..((..(((..(((......)).)..)))..))..))).))....))).))))).......---------- ( -29.00, z-score = -1.52, R) >droYak2.chr3R 27279735 115 + 28832112 AAAUGCGGCGAGCCACAGCGCUAAUCAGCGCAGGGCCAGCUGUCUCUGCACCAUCCACUAGCGCACCACCUCCUGCACCACCCAAGCCAACCACCCACACAUCGCACCACCAGCC ...(((((.(((.....((((((....(.((((((.(....).)))))).).......)))))).....))))))))..........................((.......)). ( -26.10, z-score = -0.39, R) >droSec1.super_4 5181378 93 + 6179234 AAAUGCGGCCAGCCACAGCGCUAAUCAGCGCAGGGACAGCUGUCUCUGCACCACCCACUAUCCCGCCGCCUGCACCACCGACCGCACCACCAG---------------------- ....(((((..((....))(((....)))((((((((....))))))))...............))))).(((..........))).......---------------------- ( -26.90, z-score = -1.55, R) >droSim1.chr3R 26007776 89 + 27517382 AAAUGCGGCCA----CAGCGCUAAUCAGCGCAGGGACAGCUGUCUCUGCACCACCCACUAUCCCGCCGCCUGCACCACCGACCGCACCACCAG---------------------- ....(((((..----....(((....)))((((((((....))))))))...............))))).(((..........))).......---------------------- ( -26.60, z-score = -2.03, R) >consensus AAAUGCGGCCAGCCACAGCGCUAAUCAGCGCAGGGACAGCUGUCUCUGCACCACCCACUAUCCCGCCGCCUGCACCACCGACCGCACCACCAA______________________ ....(((((..........(((....)))((((((((....))))))))...............))))).............................................. (-20.91 = -21.48 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:33 2011