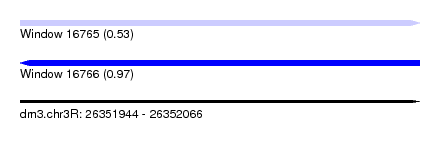
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,351,944 – 26,352,066 |
| Length | 122 |
| Max. P | 0.969543 |

| Location | 26,351,944 – 26,352,066 |
|---|---|
| Length | 122 |
| Sequences | 14 |
| Columns | 137 |
| Reading direction | forward |
| Mean pairwise identity | 68.02 |
| Shannon entropy | 0.67926 |
| G+C content | 0.48809 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -10.44 |
| Energy contribution | -11.00 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
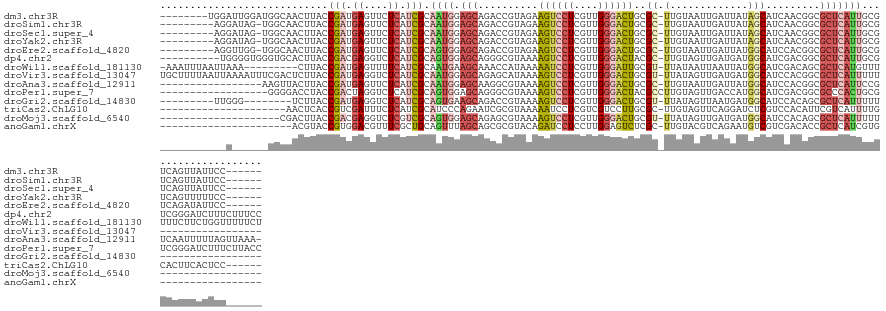
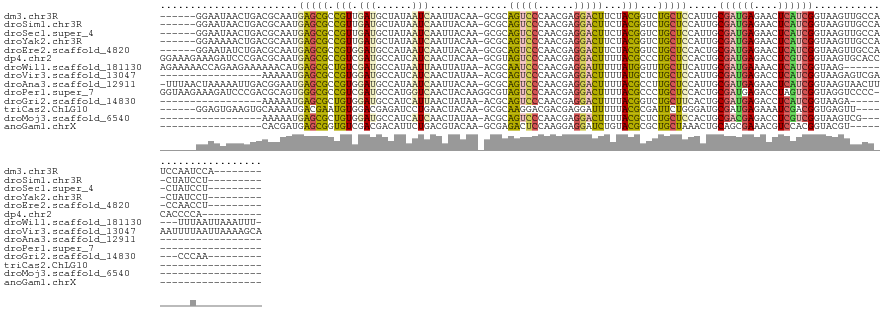
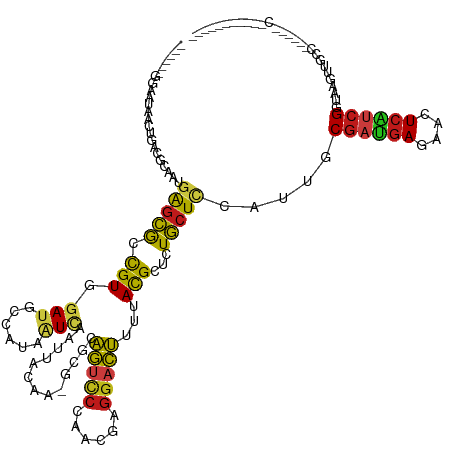
>dm3.chr3R 26351944 122 + 27905053 --------UGGAUUGGAUGGCAACUUACCGAUGAGUUCUCAUCGCAAUGGAGCAGACCGUAGAAGUCCUCGUUGGGACUGCGC-UUGUAAUUGAUUAUAGCAUCAACGGCGCUCAUUGCGUCAGUUAUUCC------ --------.(((.(((.((((........(((((....)))))((((((.(((.(.((((.((((((((....))))))..((-(.((((....))))))).)).)))))))))))))))))).))).)))------ ( -42.50, z-score = -2.52, R) >droSim1.chr3R 25997715 120 + 27517382 ---------AGGAUAG-UGGCAACUUACCGAUGAGUUCUCAUCGCAAUGGAGCAGACCGUAGAAGUCCUCGUUGGGACUGCGC-UUGUAAUUGAUUAUAGCAUCAACGGCGCUCAUUGCGUCAGUUAUUCC------ ---------.((((((-((((........(((((....)))))((((((.(((.(.((((.((((((((....))))))..((-(.((((....))))))).)).)))))))))))))))))).)))))).------ ( -42.60, z-score = -2.97, R) >droSec1.super_4 5171452 120 + 6179234 ---------AGGAUAG-UGGCAACUUACCGAUGAGUUCUCAUCGCAAUGGAGCAGACCGUAGAAGUCCUCGUUGGGACUGCGC-UUGUAAUUGAUUAUAGCAUCAACGGCGCUCAUUGCGUCAGUUAUUCC------ ---------.((((((-((((........(((((....)))))((((((.(((.(.((((.((((((((....))))))..((-(.((((....))))))).)).)))))))))))))))))).)))))).------ ( -42.60, z-score = -2.97, R) >droYak2.chr3R 27269323 120 + 28832112 ---------AGGAUAG-UGGCAACUUACCGAUGAGUUCUCAUCGCAAUGGAGCAGACCGUAGAAGUCCUCGUUGGGACUGCGC-UUGUAAUUGAUUAUAGCAUCAACGGCGCUCAUUGCGUCAGUUUUUCC------ ---------.(((.(.-((((........(((((....)))))((((((.(((.(.((((.((((((((....))))))..((-(.((((....))))))).)).)))))))))))))))))).)...)))------ ( -41.80, z-score = -2.75, R) >droEre2.scaffold_4820 8812642 120 - 10470090 ---------AGGUUGG-UGGCAACUUACCGAUGAGUUCUCAUCGCAGUGGAGCAGACCGUAGAAGUCCUCGUUGGGACUGCGC-UUGUAAUUGAUUAUGGCAUCCACGGCGCUCAUUGCGUCAGAUAUUCC------ ---------.(((.((-(....))).)))(((((....)))))((((((.(((.(.((((.((((((((....))))))..((-(.((((....))))))).)).))))))))))))))............------ ( -39.20, z-score = -0.82, R) >dp4.chr2 29611134 126 - 30794189 ----------UGGGGUGGGUGCACUUACCGACGAGGUCUCAUCGCAGUGGAGCAGGGCGUAAAAGUCCUCGUUGGGACUACGC-UUGUAGUUGAUGAUGGCAUCGACGGCGCUCAUUGCGUCGGGAUCUUUCUUUCC ----------.((((((((....))))))((.((((((((..(((((((.(((.((((((...((((((....))))))))))-))((.(((((((....))))))).))))))))))))..))))))))))...)) ( -56.40, z-score = -3.97, R) >droWil1.scaffold_181130 9850714 126 + 16660200 -AAAUUUAAUUAAA---------CUUACCGAUGAGUUUUCAUCGCAAUGAAGCAAACCAUAAAAAUCCUCGUUGGGAUUGCGU-UUAUAAUUAAUUAUGGCAUCGACAGCGCUCAUGUUUUUUCUUCUGGUUUUUCU -.............---------.....(((((......)))))....(((..((((((....((((((....))))))((((-((((((....)))))........)))))...............))))))))). ( -19.20, z-score = 0.70, R) >droVir3.scaffold_13047 15901126 119 + 19223366 UGCUUUUAAUUAAAAUUUCGACUCUUACCGAUGAGGUCUCAUCGCAAUGGAGCAGAGCAUAAAAGUCCUCGUUGGGACUGCGU-UUAUAGUUGAUGAUGGCAUCCACGGCGCUCAUUUUU----------------- ...................(.((((...((((((....))))))....))))).((((.....((((((....))))))....-.....((.((((....)))).))...))))......----------------- ( -30.00, z-score = -0.44, R) >droAna3.scaffold_12911 729694 118 - 5364042 -----------------AAGUUACUUACCGAUGAGUUCUCAUCGCAAUGGAGCAAGGCGUAAAAGUCCUCGUUGGGACUGCGC-UUGUAAUUGAUUAUGGCAUCCACGGCGCUCAUUCCGUCAAUUUUUAGUUAAA- -----------------............(((((....)))))((..((((((.((((((...((((((....))))))))))-))((((....)))).)).))))..))..........................- ( -29.30, z-score = -0.35, R) >droPer1.super_7 469119 119 - 4445127 ------------------GGGGACCUACCGACUAGGUCUCAUCGCAGUGGAGCAGGGCGUAAAAGUCCUCGUUGGGACUACGCCUUGUAGUUGACCAUGGCAUCGACGGCGCCCACUGCGUCGGGAUCUUUCUUACC ------------------.((......))((..(((((((..((((((((.(((((((((...((((((....)))))))))))))((.(((((........))))).))))))))))))..))))))).))..... ( -53.70, z-score = -3.48, R) >droGri2.scaffold_14830 2151473 102 - 6267026 ---------UUGGG--------UCUUACCGAUGAGGUCUCAUCGCAGUGAAGCAGACCGUAAAAGUCCUCGUUGGGACUGCGU-UUAUAGUUAAUGAUGGCAUCCACAGCGCUCAUUUUU----------------- ---------...((--------(((....(((((....)))))((......)))))))..((((((...((((((((.(((..-((((.....))))..)))))).)))))...))))))----------------- ( -28.00, z-score = -0.72, R) >triCas2.ChLG10 4418449 109 + 8806720 ---------------------AACUCACCGUCGAUUUCUCAUCGCAUCCCAGAAUCGCGUAAAAAUCCUCGUCGUCCUUGCGC-UUGUAGUUCAGGAUCUCGUCCACAUUCGUCAUUUUGCACUUCACUCC------ ---------------------........(.((((.....)))))....((((((.(((..........((..(((((.(.((-.....)).))))))..))........))).))))))...........------ ( -15.47, z-score = -0.26, R) >droMoj3.scaffold_6540 19418465 99 + 34148556 --------------------CGACUUACCGACGAGGUCUCGUCGCAGUGGAGCAGAGCGUAAAAGUCCUCGUUGGGACUGCGU-UUAUAGUUGAUGAUGGCAUCCACAGCGCUCAUUUUU----------------- --------------------(.(((...((((((....)))))).))).)....((((((...((((((....))))))....-.....((.((((....)))).)).))))))......----------------- ( -32.30, z-score = -1.24, R) >anoGam1.chrX 3512344 97 - 22145176 ----------------------ACGUACCGUGGACGUUUCGCUGCAGUUUAGCAGCGCGUACAGAUCCUCCUUGGAGUCUCGC-UUGUACGUCAGAAUGUCGUCGACACCGCUCAUCGUG----------------- ----------------------.......((.((((.((((((((......)))))(((((((((..(((....)))..))..-.)))))))..)))...)))).)).............----------------- ( -32.40, z-score = -1.14, R) >consensus ___________G______GGCAACUUACCGAUGAGUUCUCAUCGCAAUGGAGCAGACCGUAAAAGUCCUCGUUGGGACUGCGC_UUGUAAUUGAUUAUGGCAUCCACGGCGCUCAUUGCGUCAGUUAUUCC______ ............................(((.((....)).))).((((.(((..........((((((....))))))....................((.......))))))))).................... (-10.44 = -11.00 + 0.56)
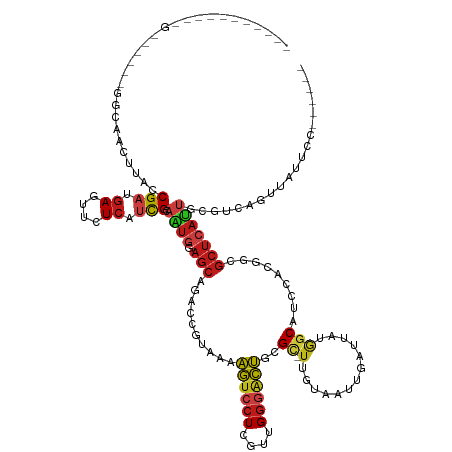
| Location | 26,351,944 – 26,352,066 |
|---|---|
| Length | 122 |
| Sequences | 14 |
| Columns | 137 |
| Reading direction | reverse |
| Mean pairwise identity | 68.02 |
| Shannon entropy | 0.67926 |
| G+C content | 0.48809 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -15.54 |
| Energy contribution | -15.04 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26351944 122 - 27905053 ------GGAAUAACUGACGCAAUGAGCGCCGUUGAUGCUAUAAUCAAUUACAA-GCGCAGUCCCAACGAGGACUUCUACGGUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAGUUGCCAUCCAAUCCA-------- ------(((.(((((.((((((((((((((((.((.(((.............)-))..(((((......))))))).)))))..))).))))))(((((....))))).)).)))))...)))......-------- ( -38.92, z-score = -2.97, R) >droSim1.chr3R 25997715 120 - 27517382 ------GGAAUAACUGACGCAAUGAGCGCCGUUGAUGCUAUAAUCAAUUACAA-GCGCAGUCCCAACGAGGACUUCUACGGUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAGUUGCCA-CUAUCCU--------- ------((..(((((.((((((((((((((((.((.(((.............)-))..(((((......))))))).)))))..))).))))))(((((....))))).)).))))))).-.......--------- ( -37.32, z-score = -2.62, R) >droSec1.super_4 5171452 120 - 6179234 ------GGAAUAACUGACGCAAUGAGCGCCGUUGAUGCUAUAAUCAAUUACAA-GCGCAGUCCCAACGAGGACUUCUACGGUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAGUUGCCA-CUAUCCU--------- ------((..(((((.((((((((((((((((.((.(((.............)-))..(((((......))))))).)))))..))).))))))(((((....))))).)).))))))).-.......--------- ( -37.32, z-score = -2.62, R) >droYak2.chr3R 27269323 120 - 28832112 ------GGAAAAACUGACGCAAUGAGCGCCGUUGAUGCUAUAAUCAAUUACAA-GCGCAGUCCCAACGAGGACUUCUACGGUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAGUUGCCA-CUAUCCU--------- ------((...((((.((((((((((((((((.((.(((.............)-))..(((((......))))))).)))))..))).))))))(((((....))))).)).)))).)).-.......--------- ( -38.42, z-score = -3.04, R) >droEre2.scaffold_4820 8812642 120 + 10470090 ------GGAAUAUCUGACGCAAUGAGCGCCGUGGAUGCCAUAAUCAAUUACAA-GCGCAGUCCCAACGAGGACUUCUACGGUCUGCUCCACUGCGAUGAGAACUCAUCGGUAAGUUGCCA-CCAACCU--------- ------((..........(((..((((((((((((((((..............-).)))((((......)))).))))))))..))))...)))(((((....)))))(((.....))).-)).....--------- ( -35.74, z-score = -1.66, R) >dp4.chr2 29611134 126 + 30794189 GGAAAGAAAGAUCCCGACGCAAUGAGCGCCGUCGAUGCCAUCAUCAACUACAA-GCGUAGUCCCAACGAGGACUUUUACGCCCUGCUCCACUGCGAUGAGACCUCGUCGGUAAGUGCACCCACCCCA---------- ((...........((((((((..((((...((.((((....)))).)).....-(((((((((......)))))...))))...))))...)))...((....)))))))...(((....))).)).---------- ( -33.50, z-score = -1.23, R) >droWil1.scaffold_181130 9850714 126 - 16660200 AGAAAAACCAGAAGAAAAAACAUGAGCGCUGUCGAUGCCAUAAUUAAUUAUAA-ACGCAAUCCCAACGAGGAUUUUUAUGGUUUGCUUCAUUGCGAUGAAAACUCAUCGGUAAG---------UUUAAUUAAAUUU- ......(((.((((...(((((((.((((....).)))))).......(((((-(...(((((......))))))))))))))).)))).....(((((....))))))))...---------.............- ( -22.80, z-score = -0.49, R) >droVir3.scaffold_13047 15901126 119 - 19223366 -----------------AAAAAUGAGCGCCGUGGAUGCCAUCAUCAACUAUAA-ACGCAGUCCCAACGAGGACUUUUAUGCUCUGCUCCAUUGCGAUGAGACCUCAUCGGUAAGAGUCGAAAUUUUAAUUAAAAGCA -----------------.(((((((((...((.((((....)))).)).((((-(...(((((......)))))))))))))).((((..(((((((((....))))).))))))))....)))))........... ( -25.60, z-score = -0.30, R) >droAna3.scaffold_12911 729694 118 + 5364042 -UUUAACUAAAAAUUGACGGAAUGAGCGCCGUGGAUGCCAUAAUCAAUUACAA-GCGCAGUCCCAACGAGGACUUUUACGCCUUGCUCCAUUGCGAUGAGAACUCAUCGGUAAGUAACUU----------------- -....(((.....((.((((........)))).))((((...........(((-(.(((((((......))))).....)))))).........(((((....)))))))))))).....----------------- ( -26.80, z-score = -0.63, R) >droPer1.super_7 469119 119 + 4445127 GGUAAGAAAGAUCCCGACGCAGUGGGCGCCGUCGAUGCCAUGGUCAACUACAAGGCGUAGUCCCAACGAGGACUUUUACGCCCUGCUCCACUGCGAUGAGACCUAGUCGGUAGGUCCCC------------------ ((............((.(((((((((((((((.......))))).........((((((((((......)))))...)))))..)).)))))))).)).((((((.....)))))))).------------------ ( -43.00, z-score = -1.62, R) >droGri2.scaffold_14830 2151473 102 + 6267026 -----------------AAAAAUGAGCGCUGUGGAUGCCAUCAUUAACUAUAA-ACGCAGUCCCAACGAGGACUUUUACGGUCUGCUUCACUGCGAUGAGACCUCAUCGGUAAGA--------CCCAA--------- -----------------...(((((..((.......))..)))))........-....(((((......))))).....(((((((......))(((((....)))))....)))--------))...--------- ( -24.30, z-score = -0.77, R) >triCas2.ChLG10 4418449 109 - 8806720 ------GGAGUGAAGUGCAAAAUGACGAAUGUGGACGAGAUCCUGAACUACAA-GCGCAAGGACGACGAGGAUUUUUACGCGAUUCUGGGAUGCGAUGAGAAAUCGACGGUGAGUU--------------------- ------(((.....((.((..((.....)).)).))....)))..((((((..-.(....)..((.(((...(((((((((.(((....)))))).)))))).))).)))).))))--------------------- ( -18.80, z-score = 1.79, R) >droMoj3.scaffold_6540 19418465 99 - 34148556 -----------------AAAAAUGAGCGCUGUGGAUGCCAUCAUCAACUAUAA-ACGCAGUCCCAACGAGGACUUUUACGCUCUGCUCCACUGCGACGAGACCUCGUCGGUAAGUCG-------------------- -----------------......(((((..(((((((....))))........-....(((((......)))))....)))..))))).(((.((((((....))))))...)))..-------------------- ( -27.30, z-score = -1.32, R) >anoGam1.chrX 3512344 97 + 22145176 -----------------CACGAUGAGCGGUGUCGACGACAUUCUGACGUACAA-GCGAGACUCCAAGGAGGAUCUGUACGCGCUGCUAAACUGCAGCGAAACGUCCACGGUACGU---------------------- -----------------....(((.((.(((..((((.........((((((.-..((..(((....)))..))))))))((((((......))))))...))))))).)).)))---------------------- ( -35.00, z-score = -1.83, R) >consensus ______GGAAUAACUGACGCAAUGAGCGCCGUGGAUGCCAUAAUCAAUUACAA_GCGCAGUCCCAACGAGGACUUUUACGCUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAGUUGCC______C___________ .......................(((((.(((.(((......))).............(((((......)))))...)))...))))).....((((((....))))))............................ (-15.54 = -15.04 + -0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:30 2011