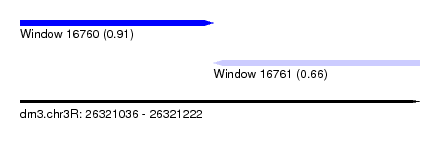
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,321,036 – 26,321,222 |
| Length | 186 |
| Max. P | 0.914011 |

| Location | 26,321,036 – 26,321,126 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 60.60 |
| Shannon entropy | 0.70947 |
| G+C content | 0.44778 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -10.08 |
| Energy contribution | -10.66 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.914011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26321036 90 + 27905053 ----CUUUAAUUAG--CCA-AAAUCGAACCAGCAGCA--GCUAGAAAUCUGCCCACCCGA---GUAGUUCGAUGUGCAUUUUUGAUUUACGUGGGCGGUUGU---- ----.......(((--((.-..........(((....--)))........((((((....---((((.((((.........)))).))))))))))))))).---- ( -21.60, z-score = -0.65, R) >droSim1.chr3R 25966311 84 + 27517382 ----CUUUAAUUAG--CCA-AAAUCGAACCAGCA-----GCUAGAAAUCUGCCCACCCGA---GU---UCGAUGUGAAUUUUUGAUUUGCGUGGGCGGUUGU---- ----..........--...-...........(((-----(((((....))((((((.(((---((---(.((........)).)))))).))))))))))))---- ( -21.10, z-score = -1.04, R) >droSec1.super_4 5140618 84 + 6179234 ----CUUUAAUUAG--CCA-AAAUCGAACCAGCA-----GCUAGAAAUCUGCCCACCCGA---GU---UCGAUGUGAAUUUUUGAUUUACGUGGGCGGUUGU---- ----..........--...-...........(((-----(((((....))((((((.((.---..---.))..(((((((...)))))))))))))))))))---- ( -20.60, z-score = -1.29, R) >droYak2.chr3R 27237807 86 + 28832112 ----CUUUAAUUAG--CCA-AAAUAUGGACAGCA-----GUUGGAAAUCUGCCCACCCGA---GU---UCGAUGCCAGUUUUUGAUUUACGUGGGCGGCUGGAU-- ----......((((--((.-.....(.(((....-----))).)......((((((..((---((---(.((........)).)))))..))))))))))))..-- ( -22.20, z-score = -0.43, R) >droEre2.scaffold_4820 8782063 86 - 10470090 ----CUUUAAUUGG--CCA-ACAAAAAGCCAGCA-----GUUGGCAGUCUGCCCACCCGAC--GUGAAUU--UUUGAAUUUUUGAUUUACGUGGGCGGCUGU---- ----.........(--(((-((............-----))))))((.((((((((.....--(((((((--...........)))))))))))))))))..---- ( -26.40, z-score = -1.58, R) >droAna3.scaffold_12911 693796 87 - 5364042 ----UUUCAAUUAC--CUACAAAACAAAAUAUUA-------AAAUCGCCCGCCCACCCGGA--GC----GGCCAUUAGUGUGGAAUUUAUGUGGGUGUUCCACCGC ----.......(((--(((((.............-------.......((((((....)).--))----))((((....))))......))))))))......... ( -22.00, z-score = -1.21, R) >droWil1.scaffold_181130 9815632 101 + 16660200 CUGUUUUUGCCCACGACUGAAAACCAAAUUGAAACCCAUUCAGCCCACAUGCUGAGUUGACUAAAAAUAUGCAAUUAAUUUAUAAUCAAUGUGGGCGCCAG----- ........((((((.........................(((((......)))))(((((.((.((((.........)))).)).))))))))))).....----- ( -21.20, z-score = -1.89, R) >consensus ____CUUUAAUUAG__CCA_AAAUCAAACCAGCA_____GCUAGAAAUCUGCCCACCCGA___GU___UCGAUGUGAAUUUUUGAUUUACGUGGGCGGUUGU____ ................................................((((((((.(((........)))..((((((.....))))))))))))))........ (-10.08 = -10.66 + 0.58)

| Location | 26,321,126 – 26,321,222 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Shannon entropy | 0.30315 |
| G+C content | 0.39227 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -15.06 |
| Energy contribution | -16.42 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26321126 96 - 27905053 GCAUAUAUUUUUCCAGAUGAAAAAUUAGACUGAAAAACAAUUGGCAGU---UGUCAGCCGGCAGGCGAUUGUGAAUUUUUCAUUUCAGCUGUCAUCUAC ..............((((((.....(((.((((((((.((((.(((((---((((........))))))))).)))).))..))))))))))))))).. ( -24.60, z-score = -2.01, R) >droSim1.chr3R 25966395 98 - 27517382 GCUUAUA-UUUUCCAGAUGAAAAACUAGAAUGAAAAACAAUUGGCAGUAGAUGUCAGUCGGAAUGCGAUUGUGAAUUUUUCAUUUCAGCUGUCAUCUAC .......-......((((((....((..(((((((((((((((.((...(((....)))....)))))))))....))))))))..))...)))))).. ( -23.50, z-score = -1.82, R) >droSec1.super_4 5140702 98 - 6179234 GCUUAUA-UUUUCCAGAUGAAAAACUAGAAUGAAAAACAAUUGGCAGUUGUUGUCAGUCGGAAUGCGAUUGUGAAUUUUUCAUUUCAGCUGUCAUCUGC .......-.....(((((((....((..((((((((...(((.((((((((..((.....))..)))))))).)))))))))))..))...))))))). ( -27.60, z-score = -2.69, R) >droYak2.chr3R 27237893 96 - 28832112 GCAUAUGCUCUGCCAGUUGAAAAAUUGGAAUGAAAAUCAAUUGGCAGU---UGGCAGUUGGCAUGCGAUGGCGAAUUUUCCAUUUUGGGCGUCAUCCAC (((..((((((((((..((...((((((........))))))..))..---))))))..)))))))(((((((.....(((.....))))))))))... ( -32.40, z-score = -2.08, R) >droEre2.scaffold_4820 8782149 99 + 10470090 GCUCGCAUUUUUCCAGUUGAAAAACUGGAAUGAAAAACAAUUGGCAAUCGUUGUCAGCCGGCCAGCGACUGCGAAUUUUUCAUUUUAGGUGUCAUCCAC ....((((((((((((((....))))))))((((((...(((.(((.((((((((....)).)))))).))).)))))))))....))))))....... ( -30.00, z-score = -2.58, R) >consensus GCUUAUA_UUUUCCAGAUGAAAAACUAGAAUGAAAAACAAUUGGCAGU_G_UGUCAGUCGGCAUGCGAUUGUGAAUUUUUCAUUUCAGCUGUCAUCUAC ...............(((((....((..((((((((...(((.(((((...((((....))))....))))).)))))))))))..))...)))))... (-15.06 = -16.42 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:26 2011