| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,308,155 – 26,308,250 |
| Length | 95 |
| Max. P | 0.890021 |

| Location | 26,308,155 – 26,308,250 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 56.01 |
| Shannon entropy | 0.61872 |
| G+C content | 0.48607 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -15.64 |
| Energy contribution | -13.09 |
| Covariance contribution | -2.54 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26308155 95 + 27905053 -GGAGACCGUAGAUUGAGUUGGCAUACCUUCCAGGUUAGGGGAAACAACUGCACAGCUGGCGCGGAGUUG-GGAGCGUAAUUCUCCUGGCUCAACAC -(....)(((....(.((((((((..(((........)))(....)...))).))))).).)))((((..-((((.......))))..))))..... ( -29.20, z-score = -0.34, R) >droSec1.super_367 1931 95 + 12961 AGCAGAGCUGGUGUUACCAAGACUU-CUUUGU-GAAAGGCCAAAAGAAUCUUAUUGCCUUGAGGCAAUCAUAGCUCGUAUCUCUGCUGGAGUAUGUC (((((((.(((.((((..((((.((-((((.(-(......))))))))))))((((((....))))))..)))).).)).))))))).......... ( -25.40, z-score = -0.53, R) >droYak2.chrU 19230801 96 + 28119190 AGCAGAGCUGGUUUUACCAAGAUUUGCAUUGCCGAAAGGCCAAGAGAAUCGUAUUGCCUUGAGGCAAUCAUAGCUCGUAUCUCUGCUGGAGUGUUC- (((((((.(((.....))).....(((...(((....)))...(((....(.((((((....)))))))....)))))).))))))).........- ( -31.40, z-score = -1.86, R) >consensus AGCAGAGCUGGUAUUACCAAGACUU_CAUUGC_GAAAGGCCAAAAGAAUCGUAUUGCCUUGAGGCAAUCAUAGCUCGUAUCUCUGCUGGAGUAUCAC .((((((.(((.....))).........((((...(((((...............)))))...)))).............))))))........... (-15.64 = -13.09 + -2.54)

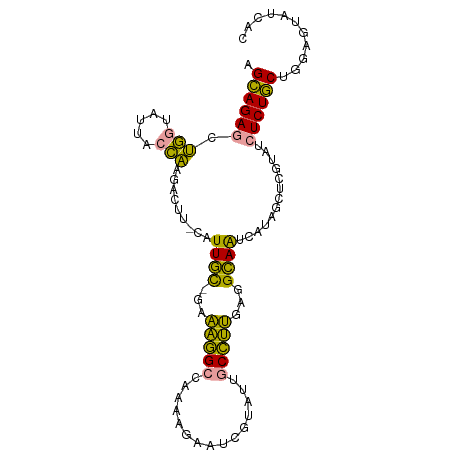
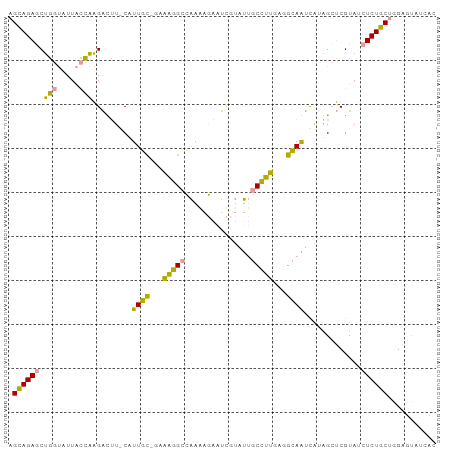
| Location | 26,308,155 – 26,308,250 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 56.01 |
| Shannon entropy | 0.61872 |
| G+C content | 0.48607 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -12.93 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.890021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26308155 95 - 27905053 GUGUUGAGCCAGGAGAAUUACGCUCC-CAACUCCGCGCCAGCUGUGCAGUUGUUUCCCCUAACCUGGAAGGUAUGCCAACUCAAUCUACGGUCUCC- ((((.(((...((((.......))))-...))).))))..((((((..((((.((((........))))((....))....)))).))))))....- ( -21.80, z-score = 0.40, R) >droSec1.super_367 1931 95 - 12961 GACAUACUCCAGCAGAGAUACGAGCUAUGAUUGCCUCAAGGCAAUAAGAUUCUUUUGGCCUUUC-ACAAAG-AAGUCUUGGUAACACCAGCUCUGCU ..........(((((((............((((((....))))))((((((..((((.......-.)))).-.))))))((.....))..))))))) ( -26.70, z-score = -2.10, R) >droYak2.chrU 19230801 96 - 28119190 -GAACACUCCAGCAGAGAUACGAGCUAUGAUUGCCUCAAGGCAAUACGAUUCUCUUGGCCUUUCGGCAAUGCAAAUCUUGGUAAAACCAGCUCUGCU -.........(((((((..........((((((((..(((((...............)))))..)))))).))....((((.....))))))))))) ( -27.26, z-score = -1.84, R) >consensus GAAAUACUCCAGCAGAGAUACGAGCUAUGAUUGCCUCAAGGCAAUACGAUUCUUUUGGCCUUUC_GCAAAG_AAGUCUUGGUAAAACCAGCUCUGCU ...........((((((............((((((....))))))..((((......((......))......)))).............)))))). (-12.93 = -13.27 + 0.34)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:22 2011