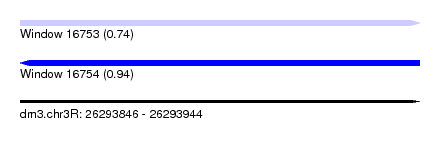
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,293,846 – 26,293,944 |
| Length | 98 |
| Max. P | 0.935948 |

| Location | 26,293,846 – 26,293,944 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 69.71 |
| Shannon entropy | 0.46158 |
| G+C content | 0.34229 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -10.44 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.736720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
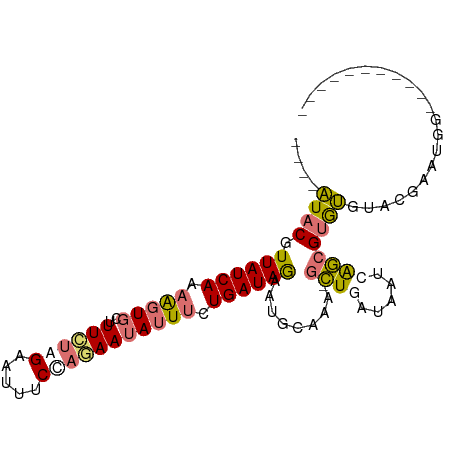
>dm3.chr3R 26293846 98 + 27905053 AUAUGUACGUUAUCAAAAGUGCUUUCUGGAAUUUCCUGAAUAUUUCUGAUAGUAUGCAAA-GCUAAUAAUCAGCGUGCGUAGCAAUGAAUGCAAACGUG .((((((((((.(((.(((((..(((.((.....)).)))))))).)))((((.......-))))......))))))))))(((.....)))....... ( -21.00, z-score = -0.73, R) >droSim1.chr3R 25939312 83 + 27517382 ----AUACGUUAUCAAAAGUGCUUUCUAGAAUUUCCAGAAUAUUUCUGAUAGAAUGCAAA-GCUGAUAAUCAGCGUGUGUACGAAUGG----------- ----(((((((((((....(((.(((((.......((((.....)))).))))).)))..-..)))))....))))))..........----------- ( -16.90, z-score = -1.04, R) >droSec1.super_4 5118183 84 + 6179234 ----AUACGUUAUCAAAAGUGCUUUCUAGAAUUUCCAAAAUAUUUCUGAUAGAAUGCAAAAGCUGAUAAUCAGCGUGUGUACGAAUGG----------- ----...(((...((....(((.((((((((............))))...)))).)))...((((.....)))).))...))).....----------- ( -14.80, z-score = -0.71, R) >droYak2.chr3R 27212581 82 + 28832112 ----AUACGUUAUCAAUGCUGCUUUGUGGAAUUUCUACAA-AUUCCUGAUAAACAGCAAACUUUUCAUACCGAUGAAUGCAAACUUA------------ ----....(((.....(((((.((((((((((((....))-)))))..)))))))))).....(((((....)))))....)))...------------ ( -15.90, z-score = -2.67, R) >consensus ____AUACGUUAUCAAAAGUGCUUUCUAGAAUUUCCAGAAUAUUUCUGAUAGAAUGCAAA_GCUGAUAAUCAGCGUGUGUACGAAUGG___________ ....((((.((((((.((((...((((((.....)))))).)))).)))))).........((((.....))))))))..................... (-10.44 = -11.12 + 0.69)

| Location | 26,293,846 – 26,293,944 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 69.71 |
| Shannon entropy | 0.46158 |
| G+C content | 0.34229 |
| Mean single sequence MFE | -17.35 |
| Consensus MFE | -10.11 |
| Energy contribution | -10.55 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
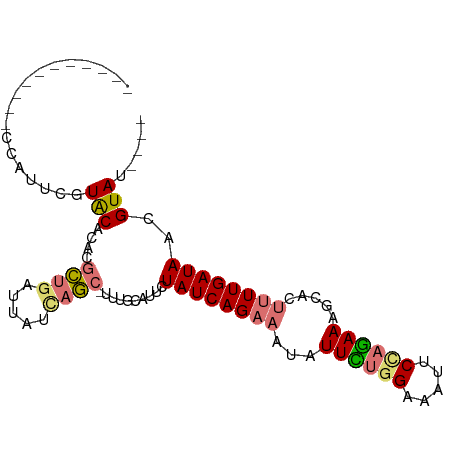
>dm3.chr3R 26293846 98 - 27905053 CACGUUUGCAUUCAUUGCUACGCACGCUGAUUAUUAGC-UUUGCAUACUAUCAGAAAUAUUCAGGAAAUUCCAGAAAGCACUUUUGAUAACGUACAUAU .((((..(((.....)))...(((.(((((...)))))-..)))....((((((((...(((.((.....)).))).....))))))))))))...... ( -17.90, z-score = -0.99, R) >droSim1.chr3R 25939312 83 - 27517382 -----------CCAUUCGUACACACGCUGAUUAUCAGC-UUUGCAUUCUAUCAGAAAUAUUCUGGAAAUUCUAGAAAGCACUUUUGAUAACGUAU---- -----------......((((....((((.....))))-.........((((((((...((((((.....)))))).....))))))))..))))---- ( -18.10, z-score = -2.72, R) >droSec1.super_4 5118183 84 - 6179234 -----------CCAUUCGUACACACGCUGAUUAUCAGCUUUUGCAUUCUAUCAGAAAUAUUUUGGAAAUUCUAGAAAGCACUUUUGAUAACGUAU---- -----------......((((....((((.....))))..........((((((((...((((((.....)))))).....))))))))..))))---- ( -15.50, z-score = -1.82, R) >droYak2.chr3R 27212581 82 - 28832112 ------------UAAGUUUGCAUUCAUCGGUAUGAAAAGUUUGCUGUUUAUCAGGAAU-UUGUAGAAAUUCCACAAAGCAGCAUUGAUAACGUAU---- ------------......(((.(((((....))))).....((((((((....(((((-((....)))))))...))))))))........))).---- ( -17.90, z-score = -1.83, R) >consensus ___________CCAUUCGUACACACGCUGAUUAUCAGC_UUUGCAUUCUAUCAGAAAUAUUCUGGAAAUUCCAGAAAGCACUUUUGAUAACGUAU____ ..................(((....((((.....))))..........((((((((...((((((.....)))))).....))))))))..)))..... (-10.11 = -10.55 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:20 2011