
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,275,100 – 26,275,190 |
| Length | 90 |
| Max. P | 0.879183 |

| Location | 26,275,100 – 26,275,190 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 66.15 |
| Shannon entropy | 0.67274 |
| G+C content | 0.50917 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -13.61 |
| Energy contribution | -14.19 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26275100 90 - 27905053 -------UAGGUG-CCCCGCAAAUUGGAACUGAUUCAAUCAGGUGCCGAAGUCUGCGGGGGCAGUUUCUUUUACGGCUCCCC--GUUUAUCA-CCCCACUA -------..((((-(((((((..((((..(((((...)))))...))))....)))))))............((((....))--))....))-))...... ( -29.60, z-score = -1.32, R) >droAna3.scaffold_12911 638744 98 + 5364042 UAGGUGCCCGGUGGCCCCA-AAAUCAACACUGAUUCAAUCAGGUGUCGAAGUCUGUGGGGGCAGUUUCUUUUAUUGGCCUCU--GUUUAUCUCAUCAAUUA .(((((..(((.((((...-......((((((((...)))).)))).((((.((((....)))))))).......)))).))--)..)))))......... ( -24.80, z-score = -0.18, R) >droEre2.scaffold_4820 8733556 79 + 10470090 -------UAGGUG-CUCCGCAAAUUGGAACUGAUUCAAUCAGGUGCCGAAGUCUGCGGGGGCAGUU----------CACCC---GUUUAUCU-GCCCAUUA -------......-..(((((..((((..(((((...)))))...))))....)))))((((((..----------.....---......))-)))).... ( -26.22, z-score = -1.38, R) >droYak2.chr3R 27193501 94 - 28832112 ---UUCGUAGGUGCCCCCGCAAAUGGGAACUGAUUCAAUCAGGUGCCGAAGUCUGCGGGGGCAGUUUCUUUUACGGCCCCC---GUUUAUCU-UCCCAUUA ---......((..((((((....))))...((((...))))))..))((((...((((((((.((.......)).))))))---))....))-))...... ( -36.20, z-score = -2.68, R) >droSec1.super_4 5099565 88 - 6179234 -------UAGGUG-CCCCGCAAAUUGGAACUGAUUCAAUCAGGUGCCGAAGCCUGC-GGGGCAGUUUCUUUUACGGCCCCC---GUUUAUCU-GCCCAUUA -------..((((-(((((((..((((..(((((...)))))...))))....)))-))))))((.......((((...))---))......-)))).... ( -30.72, z-score = -1.71, R) >droSim1.chr3R 25920666 91 - 27517382 -------UAGGUG-CCCCGCAAAUUGGAACUGAUUCAAUCAGGUGCCGAAGUCUGCGGGGGCAGUUUCUUUUACGGCCCCUC-GGUUUAUCU-GCCCAUUA -------((((((-..(((((..((((..(((((...)))))...))))....)))((((((.((.......)).)))))).-))..)))))-)....... ( -29.60, z-score = -0.71, R) >droGri2.scaffold_14830 2086055 98 + 6267026 CUCGUGCUAAAGUCUUCGACGUCACAGCAUUUAAAUUGGCGGCGGUCUUA-UCAACAAUGGCUGGUCUAGUCGAAAUAGUCGAAACUCAUU--UCCAGGUG .(((.((((.....((((((....((((.........(((....)))...-.........)))).....)))))).)))))))........--........ ( -17.60, z-score = 0.90, R) >consensus _______UAGGUG_CCCCGCAAAUUGGAACUGAUUCAAUCAGGUGCCGAAGUCUGCGGGGGCAGUUUCUUUUACGGCCCCC___GUUUAUCU_GCCCAUUA ...........((.(((((((..((((..((((.....))))...))))....))))))).))...................................... (-13.61 = -14.19 + 0.58)

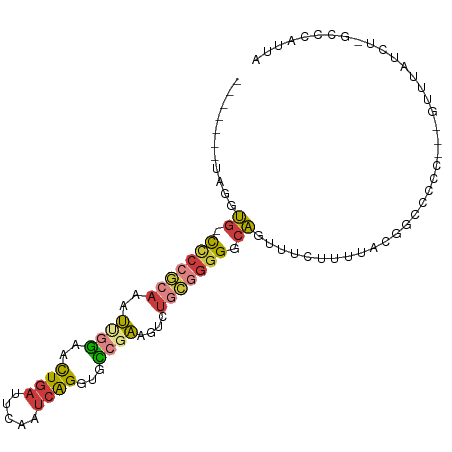
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:16 2011