
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,268,002 – 26,268,098 |
| Length | 96 |
| Max. P | 0.810530 |

| Location | 26,268,002 – 26,268,098 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Shannon entropy | 0.31613 |
| G+C content | 0.36383 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -11.74 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.751145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
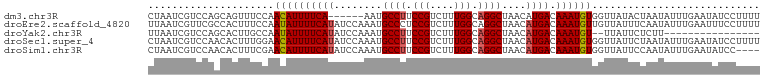
>dm3.chr3R 26268002 96 + 27905053 CUAAUCGUCCAGCAGUUUCCAACAUUUUCA------AAUGCCUUCCGUCUUUGGCAGGCUAACAUGACAAAUGUGGUUAUACUAAUAUUUGAAUAUCCUUUU ....(((......(((..(((.((((((((------...((((.(((....))).)))).....))).))))))))....)))......))).......... ( -18.00, z-score = -1.45, R) >droEre2.scaffold_4820 8726468 102 - 10470090 UUAAUCGUUCGCCACUUUCCAAUAUUUUCAUAUCCAAAUGCCCUCCGUCUUUGGCAGGCUAACAUGACAAAUGUUGUUAUUUCAAUAUUUGAAUUUCCUUUU ......(((.(((........((((....))))(((((.((.....)).)))))..))).)))....(((((((((......)))))))))........... ( -17.80, z-score = -2.51, R) >droYak2.chr3R 27186270 84 + 28832112 UUAAUCGUCCAGCACUUGCCAAUAUUUUCAUAUCCAAAUGCCUUCCGUCUUUGGCAGGCUAACAUGACAAAUGU--UUAUUCUCUU---------------- ...........((....)).(((((((((((........((((.(((....))).))))....)))).))))))--).........---------------- ( -12.60, z-score = -1.02, R) >droSec1.super_4 5092671 102 + 6179234 CUAAUCGUCCAACACUUUGGAACAUUUUCAUAUCCAAAUGCCUUCCGUCUUUGGCAGGCUAACAUGACAAAUGUGGUUAUUCUAAUAUUUGAAUAUCCUUUU .......(((((....)))))((((((((((........((((.(((....))).))))....)))).))))))((.(((((........))))).)).... ( -23.00, z-score = -2.96, R) >droSim1.chr3R 25913909 98 + 27517382 CUAAUCGUCCAACACUUUCGAACAUUUUCAUAUCCAAAUGCCUUCCGUCUUUGGCAGGCUAACAUGACAAAUGUGGUUAUUCCAAUAUUUGAAUAUCC---- ....(((...........)))((((((((((........((((.(((....))).))))....)))).))))))((.(((((........))))).))---- ( -17.10, z-score = -1.36, R) >consensus CUAAUCGUCCAACACUUUCCAACAUUUUCAUAUCCAAAUGCCUUCCGUCUUUGGCAGGCUAACAUGACAAAUGUGGUUAUUCUAAUAUUUGAAUAUCCUUUU .....................((((((((((........((((.(((....))).))))....)))).))))))............................ (-11.74 = -11.90 + 0.16)
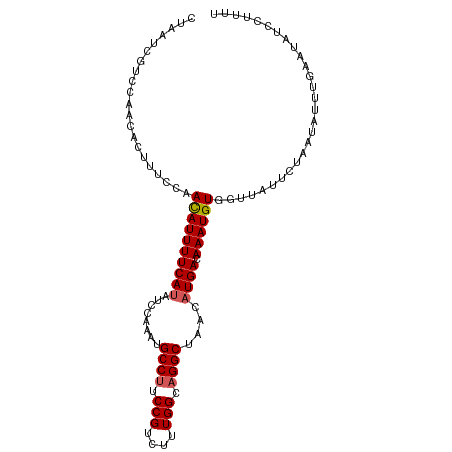
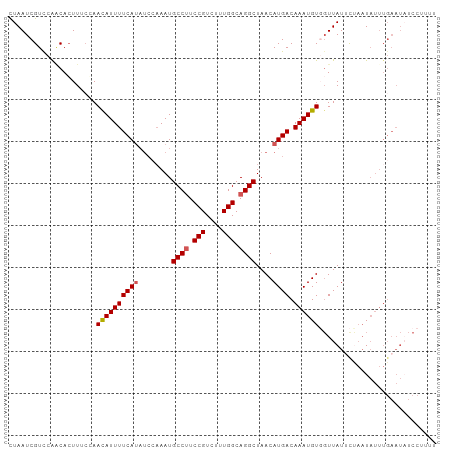
| Location | 26,268,002 – 26,268,098 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.09 |
| Shannon entropy | 0.31613 |
| G+C content | 0.36383 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.90 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26268002 96 - 27905053 AAAAGGAUAUUCAAAUAUUAGUAUAACCACAUUUGUCAUGUUAGCCUGCCAAAGACGGAAGGCAUU------UGAAAAUGUUGGAAACUGCUGGACGAUUAG .................((((((...((((((((.(((.....((((.((......)).))))...------)))))))).)))....))))))........ ( -18.50, z-score = -0.60, R) >droEre2.scaffold_4820 8726468 102 + 10470090 AAAAGGAAAUUCAAAUAUUGAAAUAACAACAUUUGUCAUGUUAGCCUGCCAAAGACGGAGGGCAUUUGGAUAUGAAAAUAUUGGAAAGUGGCGAACGAUUAA .........((((.....)))).......(.((((((((....((((.((......)).)))).(((.(((((....))))).))).)))))))).)..... ( -22.50, z-score = -2.29, R) >droYak2.chr3R 27186270 84 - 28832112 ----------------AAGAGAAUAA--ACAUUUGUCAUGUUAGCCUGCCAAAGACGGAAGGCAUUUGGAUAUGAAAAUAUUGGCAAGUGCUGGACGAUUAA ----------------..........--.((((((((......((((.((......)).)))).....(((((....)))))))))))))............ ( -20.80, z-score = -2.28, R) >droSec1.super_4 5092671 102 - 6179234 AAAAGGAUAUUCAAAUAUUAGAAUAACCACAUUUGUCAUGUUAGCCUGCCAAAGACGGAAGGCAUUUGGAUAUGAAAAUGUUCCAAAGUGUUGGACGAUUAG ....((.(((((........))))).))....(((((......((((.((......)).)))).(((((((((....))).))))))......))))).... ( -26.60, z-score = -3.02, R) >droSim1.chr3R 25913909 98 - 27517382 ----GGAUAUUCAAAUAUUGGAAUAACCACAUUUGUCAUGUUAGCCUGCCAAAGACGGAAGGCAUUUGGAUAUGAAAAUGUUCGAAAGUGUUGGACGAUUAG ----((.(((((........))))).))....(((((......((((.((......)).)))).(((((((((....))))))))).......))))).... ( -26.90, z-score = -2.85, R) >consensus AAAAGGAUAUUCAAAUAUUAGAAUAACCACAUUUGUCAUGUUAGCCUGCCAAAGACGGAAGGCAUUUGGAUAUGAAAAUGUUGGAAAGUGCUGGACGAUUAG ................................(((((......((((.((......)).)))).(((.(((((....))))).))).......))))).... (-13.62 = -14.90 + 1.28)


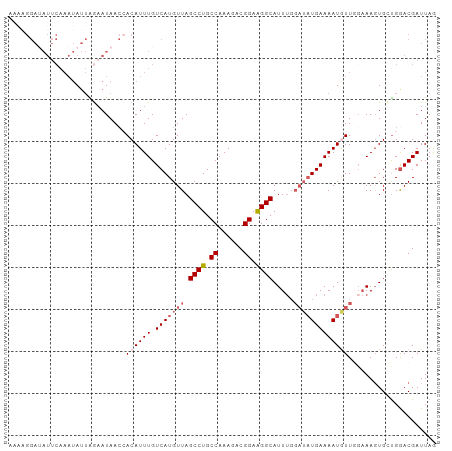
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:15 2011