| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,233,022 – 26,233,076 |
| Length | 54 |
| Max. P | 0.931301 |

| Location | 26,233,022 – 26,233,076 |
|---|---|
| Length | 54 |
| Sequences | 12 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 70.37 |
| Shannon entropy | 0.56217 |
| G+C content | 0.38289 |
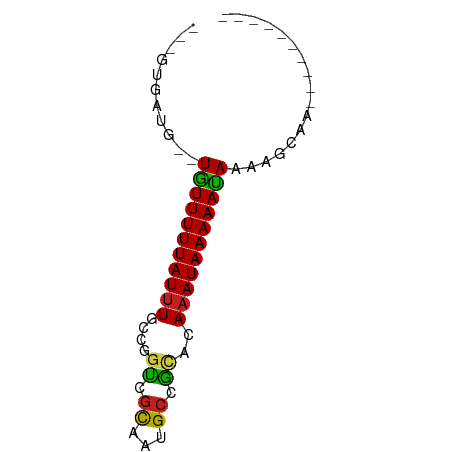
| Mean single sequence MFE | -13.21 |
| Consensus MFE | -5.88 |
| Energy contribution | -5.32 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
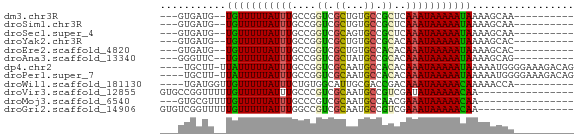
>dm3.chr3R 26233022 54 - 27905053 ---GUGAUG--UGUUUUUAUUUGCCGGUCGCUGUGCCGCUCAAAUAAAAAUAAAAGCAA---------- ---((....--((((((((((((.((((......))))..))))))))))))...))..---------- ( -15.20, z-score = -2.99, R) >droSim1.chr3R 25879473 54 - 27517382 ---GUGAUG--UGUUUUUAUUUGCCGGUCGCUGUGCCGCUCAAAUAAAAAUAAAAGCAA---------- ---((....--((((((((((((.((((......))))..))))))))))))...))..---------- ( -15.20, z-score = -2.99, R) >droSec1.super_4 5057741 54 - 6179234 ---GUGAUG--UGUUUUUAUUUGCCGGUCGCAGUGCCGCUCAAAUAAAAAUAAAAGCAA---------- ---((....--((((((((((((.((((......))))..))))))))))))...))..---------- ( -15.20, z-score = -2.78, R) >droYak2.chr3R 27150051 54 - 28832112 ---GUGAUG--UGUUUUUAUUUGCCGGUCGCUGUGCCGCACAAAUAAAAAUAAAAGCAC---------- ---(((...--((((((((((((.((((......))))..))))))))))))....)))---------- ( -16.00, z-score = -2.62, R) >droEre2.scaffold_4820 8689618 54 + 10470090 ---GUGAUG--UGUUUUUAUUUGCCGGUCGCUGUGCCACACAAAUAAAAAUAAAAGCAC---------- ---(((...--((((((((((((..(((......)))...))))))))))))....)))---------- ( -13.50, z-score = -1.84, R) >droAna3.scaffold_13340 21251830 54 - 23697760 ---GGGUUC--UGUUUUUAUUUGCCGGUCGCUAUGCCGCACAAAUAAAAAUAAAAGCAG---------- ---.(.((.--((((((((((((.((((......))))..)))))))))))).)).)..---------- ( -14.80, z-score = -2.35, R) >dp4.chr2 29501732 64 + 30794189 ----UGCUU-UUAUUUUUAUUUGCCGGUCGCAAUGCCACACAAAUAAAAAUAAAAAUGGGGAAAGACAG ----...((-(((((((((((((..(((......)))...))))))))))))))).............. ( -13.50, z-score = -1.88, R) >droPer1.super_7 352108 64 + 4445127 ----UGCUU-UUAUUUUUAUUUGCCGGUCGCAAUGCCACACAAAUAAAAAUAAAAAUGGGGAAAGACAG ----...((-(((((((((((((..(((......)))...))))))))))))))).............. ( -13.50, z-score = -1.88, R) >droWil1.scaffold_181130 9730835 55 - 16660200 ----UGAUGGUUGUUUUUAUUUCUGUGGCAUUGCGACCGACAAAUAAAAACAAAAACCA---------- ----...((((((((((((((((.((.((...)).)).))..))))))))))...))))---------- ( -13.50, z-score = -2.34, R) >droVir3.scaffold_12855 3342423 53 - 10161210 GUGCCGGUUUUUGUUUUUAUUUGCCCGUCGCAAUGCCGUCGAUAUAAAAACAA---------------- ..........((((((((((.....((.((......)).))..))))))))))---------------- ( -8.00, z-score = 0.23, R) >droMoj3.scaffold_6540 24302920 50 + 34148556 ---GUGCGUUUUGUUUUUAUUUGCCCGUCGCAAUGCCAACGAAAUAAAAACAA---------------- ---.......((((((((((((...(((.((...))..)))))))))))))))---------------- ( -8.30, z-score = -0.38, R) >droGri2.scaffold_14906 6033753 53 - 14172833 GUGUCGGUUUUUGUUUUUAUUUGGCCGUCGCAAUGCCGUCGAAAUAAAAACAA---------------- ......(((((((((((....((((.........))))..)))))))))))..---------------- ( -11.80, z-score = -1.11, R) >consensus ___GUGAUG__UGUUUUUAUUUGCCGGUCGCAAUGCCGCACAAAUAAAAAUAAAAGCAA__________ ...........(((((((((((....((.((...)).))..)))))))))))................. ( -5.88 = -5.32 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:13 2011