
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,219,624 – 26,219,707 |
| Length | 83 |
| Max. P | 0.696791 |

| Location | 26,219,624 – 26,219,707 |
|---|---|
| Length | 83 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 70.81 |
| Shannon entropy | 0.54474 |
| G+C content | 0.39988 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -11.61 |
| Energy contribution | -12.36 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
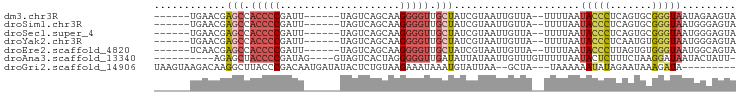
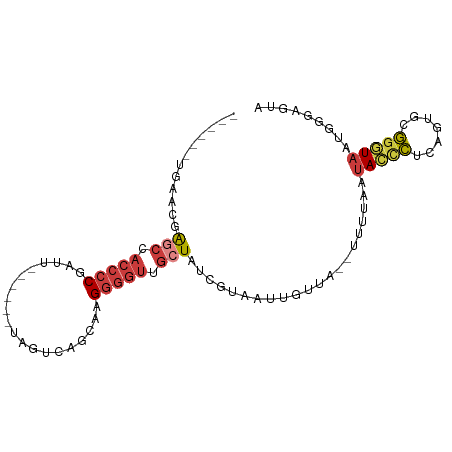
>dm3.chr3R 26219624 83 + 27905053 ------UGAACGAGCCACCCCGAUU------UAGUCAGCAAGGGGUUGCUAUCGUAAUUGUUA--UUUUAAUACCCUCAGUGCGGGUAAUAGAAGUA ------...((((((.(((((....------..........))))).))..))))........--(((((.(((((.......))))).)))))... ( -22.54, z-score = -1.68, R) >droSim1.chr3R 25873925 83 + 27517382 ------UGAACGAGCCACCCCGAUU------UAGUCAGCAAGGGGUUGCUAUCGUAAUUGUUA--UUUUAAUACCCUCAGUGCGGGUAAUGGGAGUA ------...((((((.(((((....------..........))))).))..))))........--(((((.(((((.......))))).)))))... ( -21.74, z-score = -0.49, R) >droSec1.super_4 5052540 83 + 6179234 ------UGAACGAGCCACCCCGAUU------UAGUCAGCAAGGGGUUGCUAUCGUAAUUGUUA--UUUUAAUACCCUCAGUGCGGGUAAUGGGAGUA ------...((((((.(((((....------..........))))).))..))))........--(((((.(((((.......))))).)))))... ( -21.74, z-score = -0.49, R) >droYak2.chr3R 27144648 83 + 28832112 ------UGAACGAGCCACCCCGAUU------UAGUCAGCAAGGGGUUGCUAUCGUAAUUGUUA--UUUUAAUACCCUCAAUGUGGGUAAUGGGAGUA ------...((((((.(((((....------..........))))).))..))))........--(((((.(((((.......))))).)))))... ( -22.34, z-score = -0.93, R) >droEre2.scaffold_4820 8681843 83 - 10470090 ------UCAACGAGCCACCCCGAUU------UAGUCAGCAAGGGGUUGCUAUCGUAAUUGUUA--UUUUAAUACCCUUAGUGUGGGUAAUGGCAGUA ------...((((((.(((((....------..........))))).))..)))).(((((((--(.....(((((.......))))))))))))). ( -24.84, z-score = -2.04, R) >droAna3.scaffold_13340 21246446 82 + 23697760 ----------AGAGCUACCCCGAUAG----GUAGUCACUAGGGGGUUGAUAUUAUAAUUGUUUGUUUUUAAUACUCUUUCUAAGGAUAAUACUAUU- ----------.((.(((((......)----))))))..(((..((..(.(((((.((........)).)))))).))..)))..............- ( -13.30, z-score = 1.05, R) >droGri2.scaffold_14906 6026691 83 + 14172833 UAAGUAAGACAAGGCUUACCCGACAAUGAUAUACUCUGUAAGAAAUAAAUGUAUUAA--GCUA---UAAAAAAUAUAGAAUAAAGAUA--------- ...(((((......))))).......(((((((...(((.....)))..))))))).--.(((---((.....)))))..........--------- ( -6.50, z-score = 0.18, R) >consensus ______UGAACGAGCCACCCCGAUU______UAGUCAGCAAGGGGUUGCUAUCGUAAUUGUUA__UUUUAAUACCCUCAGUGCGGGUAAUGGGAGUA ............(((.(((((....................))))).))).....................(((((.......)))))......... (-11.61 = -12.36 + 0.76)

| Location | 26,219,624 – 26,219,707 |
|---|---|
| Length | 83 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 70.81 |
| Shannon entropy | 0.54474 |
| G+C content | 0.39988 |
| Mean single sequence MFE | -16.43 |
| Consensus MFE | -10.80 |
| Energy contribution | -11.01 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.46 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
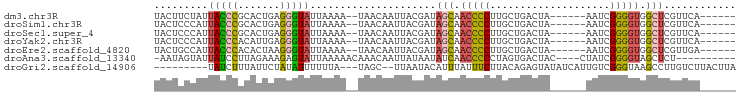
>dm3.chr3R 26219624 83 - 27905053 UACUUCUAUUACCCGCACUGAGGGUAUUAAAA--UAACAAUUACGAUAGCAACCCCUUGCUGACUA------AAUCGGGGUGGCUCGUUCA------ ......((.(((((.......))))).))...--........((((..((.(((((..........------....))))).))))))...------ ( -19.34, z-score = -1.18, R) >droSim1.chr3R 25873925 83 - 27517382 UACUCCCAUUACCCGCACUGAGGGUAUUAAAA--UAACAAUUACGAUAGCAACCCCUUGCUGACUA------AAUCGGGGUGGCUCGUUCA------ .........(((((.......)))))......--........((((..((.(((((..........------....))))).))))))...------ ( -19.14, z-score = -0.74, R) >droSec1.super_4 5052540 83 - 6179234 UACUCCCAUUACCCGCACUGAGGGUAUUAAAA--UAACAAUUACGAUAGCAACCCCUUGCUGACUA------AAUCGGGGUGGCUCGUUCA------ .........(((((.......)))))......--........((((..((.(((((..........------....))))).))))))...------ ( -19.14, z-score = -0.74, R) >droYak2.chr3R 27144648 83 - 28832112 UACUCCCAUUACCCACAUUGAGGGUAUUAAAA--UAACAAUUACGAUAGCAACCCCUUGCUGACUA------AAUCGGGGUGGCUCGUUCA------ .........(((((.......)))))......--........((((..((.(((((..........------....))))).))))))...------ ( -18.44, z-score = -0.78, R) >droEre2.scaffold_4820 8681843 83 + 10470090 UACUGCCAUUACCCACACUAAGGGUAUUAAAA--UAACAAUUACGAUAGCAACCCCUUGCUGACUA------AAUCGGGGUGGCUCGUUGA------ .........(((((.......)))))......--.........(((((((.(((((..........------....))))).))).)))).------ ( -19.94, z-score = -1.26, R) >droAna3.scaffold_13340 21246446 82 - 23697760 -AAUAGUAUUAUCCUUAGAAAGAGUAUUAAAAACAAACAAUUAUAAUAUCAACCCCCUAGUGACUAC----CUAUCGGGGUAGCUCU---------- -...................(((((((((.((........)).))))....(((((.(((.......----)))..))))).)))))---------- ( -10.80, z-score = 0.14, R) >droGri2.scaffold_14906 6026691 83 - 14172833 ---------UAUCUUUAUUCUAUAUUUUUUA---UAGC--UUAAUACAUUUAUUUCUUACAGAGUAUAUCAUUGUCGGGUAAGCCUUGUCUUACUUA ---------.....(((..(((((.....))---))).--.))).................(((((...((.....(((....)))))...))))). ( -8.20, z-score = -0.06, R) >consensus UACUCCCAUUACCCGCACUGAGGGUAUUAAAA__UAACAAUUACGAUAGCAACCCCUUGCUGACUA______AAUCGGGGUGGCUCGUUCA______ .........(((((.......))))).....................(((.(((((....................))))).)))............ (-10.80 = -11.01 + 0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:11 2011