| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,201,573 – 26,201,708 |
| Length | 135 |
| Max. P | 0.999425 |

| Location | 26,201,573 – 26,201,679 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 56.31 |
| Shannon entropy | 0.74819 |
| G+C content | 0.47787 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.70 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26201573 106 + 27905053 --UAAGCCUAGAUAUAUAAGAUAUACAUAAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUU-AGCCU----AAGUCCAAAUAUAUAAAAAACUGUA --..........((((((................((.(((.(((((((((.....))))))))).)))...))(((((-.....----.)))))...)))))).......... ( -29.90, z-score = -1.65, R) >droSim1.chr3h_random 862046 96 + 1452968 --UAACUAUAUAUAUGUAGGAAACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUGGACUUAGGCUA----AGAAACCAUUGUAU----------- --........((..(.(((((.((((......)))).((.(((((((.(((...))).))))))).))))))).)..))((...----.....)).......----------- ( -32.10, z-score = -1.43, R) >droSec1.super_16 19895 96 + 1878335 --AAUAUAUAUAUAUGUAGGAAACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUA----AGAAACCAUUGUAU----------- --.(((((......((.((((.((((......)))).((.(((((((.(((...))).))))))).)))))))).....((...----.....))..)))))----------- ( -31.10, z-score = -1.36, R) >droEre2.scaffold_4855 234970 96 - 242605 ------------UA-AGCACAGAUAUAUAAGAGAGCCGCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUU----UAGCCGAAAUAUACUUAAGAAUGUA ------------..-...(((......((((((((((((..((((((..((...))..)))))).......)))))))).((..----...))........))))....))). ( -29.20, z-score = -0.22, R) >droPer1.super_81 10284 106 + 277874 UACUAACAUAUAAGGGAUAAGAACACAUAGUUCUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUUGGCUUACAAAAGCCCAAAAAAUUCAG------- ..............((...(((((.....))))).))(((.(((((((((.....))))))))).)))...(.((((((((.....)))))))))...........------- ( -40.80, z-score = -4.30, R) >consensus __UAAACAUAUAUAUGUAAGAAACACAUAAAAGCGCCGCUCCGCUGCAGGCGUAGCCGGCAGCGCAGCUCCGCGGCUUAAGCUU____AAAACCCAAUAUAU__A________ ..................................((((((.(((((((((.....))))))))).))).....)))..................................... (-19.62 = -19.70 + 0.08)


| Location | 26,201,573 – 26,201,679 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 56.31 |
| Shannon entropy | 0.74819 |
| G+C content | 0.47787 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26201573 106 - 27905053 UACAGUUUUUUAUAUAUUUGGACUU----AGGCU-AAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGUUCUUAUGUAUAUCUUAUAUAUCUAGGCUUA-- ..........(((((((..((((..----..((.-.....))((.(((.((((((.((.....)).)))))).))).))))))..)))))))...................-- ( -32.20, z-score = -0.55, R) >droSim1.chr3h_random 862046 96 - 1452968 -----------AUACAAUGGUUUCU----UAGCCUAAGUCCAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUUUCCUACAUAUAUAUAGUUA-- -----------.......((((...----.)))).......((((((.(((((((.(((...))).))))))).))(((((......)))))))))...............-- ( -34.40, z-score = -2.72, R) >droSec1.super_16 19895 96 - 1878335 -----------AUACAAUGGUUUCU----UAGCCUAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUUUCCUACAUAUAUAUAUAUU-- -----------.......((((...----.)))).......((((((.(((((((.(((...))).))))))).))(((((......)))))))))...............-- ( -34.50, z-score = -2.77, R) >droEre2.scaffold_4855 234970 96 + 242605 UACAUUCUUAAGUAUAUUUCGGCUA----AACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGCGGCUCUCUUAUAUAUCUGUGCU-UA------------ ........((((((((....((...----..)).......((...(((.((((((..((...))..)))))).))).)).............))))))-))------------ ( -25.60, z-score = 0.00, R) >droPer1.super_81 10284 106 - 277874 -------CUGAAUUUUUUGGGCUUUUGUAAGCCAAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAGAACUAUGUGUUCUUAUCCCUUAUAUGUUAGUA -------((((........((((((((.....))))))))..((((((.(((((((.(.....).))))))).)))...(((((.....)))))..)))........)))).. ( -39.60, z-score = -1.94, R) >consensus ________U__AUAUAUUGGGUUUU____AAGCCAAAGCCGCGGAGCUGCGCUGCCGGCAACGCCAGCAGCGGAGCGGCGCUCAUAUGUGUUUCUUACAUAUAUAUAUUUA__ ..........................................(..(((.((((((.((.....)).)))))).)))..).................................. (-18.18 = -18.98 + 0.80)


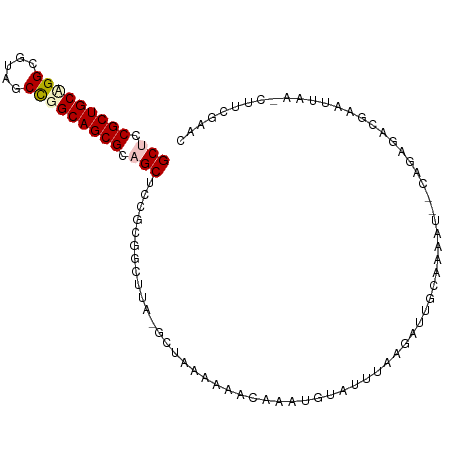
| Location | 26,201,608 – 26,201,708 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 59.92 |
| Shannon entropy | 0.71647 |
| G+C content | 0.49166 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -18.70 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26201608 100 + 27905053 GCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUUA-GCCUAAGUCCAAAUAUAUAAAAAACUGUAAAAUCGGAGAGACUCUGUAGACGUUGAGC (((.(((((((((.....))))))))).)))...(((((((.-.....)))))............(((.((.....(((((...)))))...)))))..)) ( -31.20, z-score = -1.50, R) >droSim1.chr3h_random 862081 97 + 1452968 GCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUGGACUUAGGCUAAGAAACCAUUGUAUUUA-GAUCGCCAAAU--GAUUGACGAAUUAA-CUUCGAAU ((.(((((((.(((...))).))))))).)).((((....))))...........(((...(((-(.(((.(((..--..))).))).))))-...))).. ( -27.40, z-score = -0.84, R) >droSec1.super_16 19930 97 + 1878335 GCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUAAGAAACCAUUGUAUUUA-GAUCGCCAAAU--CAUUGACGAAUUAA-CUUCGAAU ((.(((((((.(((...))).))))))).))...((((....((........)).......(((-(.(((.(((..--..))).))).))))-..)))).. ( -28.90, z-score = -1.58, R) >droEre2.scaffold_4855 234994 99 - 242605 GCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCCGAAAUAUACUUAAGAAUGUAUCGC--CAGAUACUUGUGAAUGUUUGAAC (((.((((((..((...))..)))))).)))..(((((((((((((.((.......)).))))))))..(((((..--..))))))))))........... ( -33.20, z-score = -1.59, R) >droPer1.super_81 10321 89 + 277874 GCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUU---UUGGCUUACAAAAGCCCAAAAAAUU-CAGAAU-AUAAAAACCAAUAAAAC------- (((.(((((((((.....))))))))).)))...(.(((((---(((.....)))))))))........-......-.................------- ( -33.20, z-score = -4.61, R) >consensus GCUCCGCUGCAGGCGUAGCCGGCAGCGCAGCUCCGCGGCUUA_GCUAAAAAACAAAUGUAUUUAAGAUUGCAAAAU__CAGAGACGAAUUAA_CUUCGAAC (((.(((((((((.....))))))))).)))...................................................................... (-18.70 = -19.18 + 0.48)
| Location | 26,201,608 – 26,201,708 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 59.92 |
| Shannon entropy | 0.71647 |
| G+C content | 0.49166 |
| Mean single sequence MFE | -30.81 |
| Consensus MFE | -16.22 |
| Energy contribution | -17.70 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
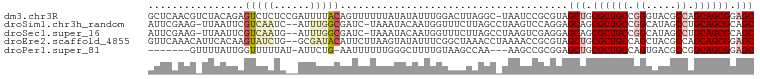
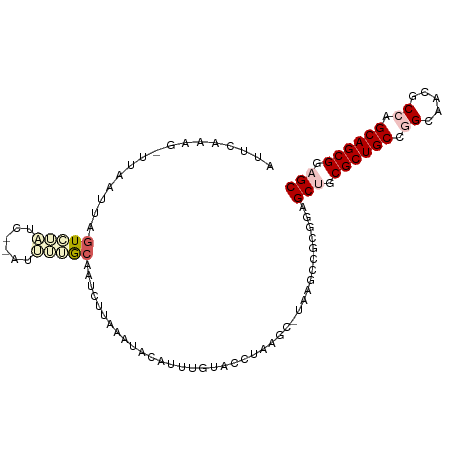
>dm3.chr3R 26201608 100 - 27905053 GCUCAACGUCUACAGAGUCUCUCCGAUUUUACAGUUUUUUAUAUAUUUGGACUUAGGC-UAAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGC .....((((......(((((.(((((....................)))))...))))-).....)))).(((.((((((.((.....)).)))))).))) ( -29.35, z-score = -0.67, R) >droSim1.chr3h_random 862081 97 - 1452968 AUUCGAAG-UUAAUUCGUCAAUC--AUUUGGCGAUC-UAAAUACAAUGGUUUCUUAGCCUAAGUCCAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGC ........-(((..(((((((..--..)))))))..-)))........((((((..((....))..)))))).(((((((.(((...))).)))))))... ( -33.10, z-score = -2.67, R) >droSec1.super_16 19930 97 - 1878335 AUUCGAAG-UUAAUUCGUCAAUG--AUUUGGCGAUC-UAAAUACAAUGGUUUCUUAGCCUAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGC ........-(((..(((((((..--..)))))))..-)))........((((((..((....))..)))))).(((((((.(((...))).)))))))... ( -35.90, z-score = -2.89, R) >droEre2.scaffold_4855 234994 99 + 242605 GUUCAAACAUUCACAAGUAUCUG--GCGAUACAUUCUUAAGUAUAUUUCGGCUAAACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGC ((((............(((((..--..)))))................((((((..(........)..))))))((((((..((...))..)))))))))) ( -23.90, z-score = -0.20, R) >droPer1.super_81 10321 89 - 277874 -------GUUUUAUUGGUUUUUAU-AUUCUG-AAUUUUUUGGGCUUUUGUAAGCCAA---AAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGC -------.................-......-.....((((((((((((.....)))---))))).))))(((.(((((((.(.....).))))))).))) ( -31.80, z-score = -1.68, R) >consensus AUUCAAAG_UUAAUUAGUCUAUC__AUUUUGCAAUCUUAAAUACAUUUGUACCUAAGC_UAAGCCGCGGAGCUGCGCUGCCGGCAACGCCAGCAGCGGAGC ................(((((......)))))......................................(((.((((((.((.....)).)))))).))) (-16.22 = -17.70 + 1.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:09 2011