| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,199,145 – 26,199,258 |
| Length | 113 |
| Max. P | 0.926426 |

| Location | 26,199,145 – 26,199,258 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.63 |
| Shannon entropy | 0.53462 |
| G+C content | 0.49920 |
| Mean single sequence MFE | -32.76 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
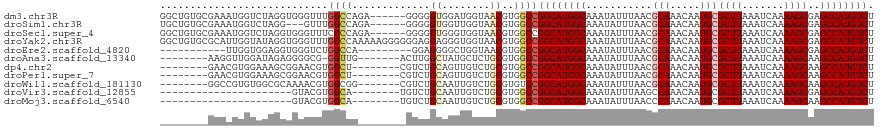
>dm3.chr3R 26199145 113 + 27905053 GGCUGUGCGAAAUGGUCUAGGUGGGUUUGGCCAGA------GGGGGUGGAUGGUAACGUGGCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCGAGCCAUGUCU ((((..(((..((.(((((....((.....))...------.....))))).))..)))))))((((((((...........(((.....)))((((.......))))..)))))))). ( -33.80, z-score = -0.18, R) >droSim1.chr3R 25860213 110 + 27517382 UGCUGUGCGAAAUGGUCUAGG---GUUUGGCCAGA------GGGGGUGGUUGGUAACGUGGCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCGAGCCAUGUCU .((..(.(....(((((....---....)))))..------..).)..)).(((......)))((((((((...........(((.....)))((((.......))))..)))))))). ( -33.00, z-score = -0.13, R) >droSec1.super_4 5039695 113 + 6179234 GGCUGUGCGAAAUGGUCUAGGUGGGUUUCGCCAGA------GGGGGUGGGUGGUAACGUGGCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCGAGCCAUGUCU ..(((.(((((((..((.....)))))))))))).------..((.((.((....)).)).))((((((((...........(((.....)))((((.......))))..)))))))). ( -35.20, z-score = -0.40, R) >droYak2.chr3R 27131247 119 + 28832112 GGCUGUGCGCAUUGGUAUAGGUGGGUUUGGCCAAAAAGGGGGGAGGAGGGUGGUAACGUGGCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCGAGCCAUGUCU (((((.((.((((......)))).)).)))))............((.(.((....)).)..))((((((((...........(((.....)))((((.......))))..)))))))). ( -35.10, z-score = -0.35, R) >droEre2.scaffold_4820 8668702 99 - 10470090 -----------UUGGUGGAGGUGGGUCUGGCCA---------GGAGGGGCUGGUAACGUGGCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCGAGCCAUGUCU -----------..(((.(..((....(..(((.---------.....)))..)..)).).)))((((((((...........(((.....)))((((.......))))..)))))))). ( -33.50, z-score = -1.31, R) >droAna3.scaffold_13340 21231587 103 + 23697760 --------AAGGUUGGAUAGAGGGGCG-GGUUG-------ACUUGGCUAUGCUCUGCGUGGCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCAAGCCAUGUCU --------.((((..(...(.....).-..)..-------))))((((((((...))))))))((((((((...........(((.....)))((((.......))))..)))))))). ( -35.50, z-score = -1.88, R) >dp4.chr2 29484765 103 - 30794189 --------GAACGUGGAAAGCGGAACGUGGCU--------CGUCUGCAGUUGUCUGCGUGGCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCAAGCCAUGUCU --------..(((..((..(((((.((.....--------))))))).....))..)))....((((((((...........(((.....)))((((.......))))..)))))))). ( -34.60, z-score = -1.63, R) >droPer1.super_7 335017 103 - 4445127 --------GAACGUGGAAAGCGGAACGUGGCU--------CGUCUGCAGUUGUCUGCGUGGCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCAAGCCAUGUCU --------..(((..((..(((((.((.....--------))))))).....))..)))....((((((((...........(((.....)))((((.......))))..)))))))). ( -34.60, z-score = -1.63, R) >droWil1.scaffold_181130 9701006 104 + 16660200 --------GGCCGUGUGGCGCAAAACGUGGCGG-------CGUCUGCAAUUGUCUGCGUGUCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCAAGCCAUGUCU --------.(((((((........)))))))((-------((..((((......)))))))).((((((((...........(((.....)))((((.......))))..)))))))). ( -32.60, z-score = 0.05, R) >droVir3.scaffold_12855 3321505 89 + 10161210 ----------------------GUACGUGGCA--------UGUCUGCAAUUGUCUGCGUGGCCGGCAUGGCAAAUAUUUAAGCCAAACAAUGCGCUUAAAUCAAAAGCGAGCCAUGUCU ----------------------......((((--------(((..((....))..)))).)))((((((((.....(((((((((.....)).)))))))((......)))))))))). ( -28.60, z-score = -1.48, R) >droMoj3.scaffold_6540 24280144 89 - 34148556 ----------------------GUACGUGGCA--------UGUCUGCAAUUGUCUGCGUGGCCGGCAUGGCAAAUAUUUAACCCAAACAAUGCGCUUAAAUCAAAAGCAAGCCAUGUCU ----------------------..(((((((.--------.....(((.((((.((.((.(((.....))).........)).)).)))))))((((.......))))..))))))).. ( -23.90, z-score = -0.47, R) >consensus ________GAAAUGGUAUAGGUGGGCGUGGCCA________GUCGGCAGUUGUCUGCGUGGCCGGCAUGGCAAAUAUUUAACGCAAACAAUGCGCUUAAAUCAAAAGCGAGCCAUGUCU ............................((((...........................))))((((((((...........(((.....)))((((.......))))..)))))))). (-21.24 = -21.68 + 0.44)
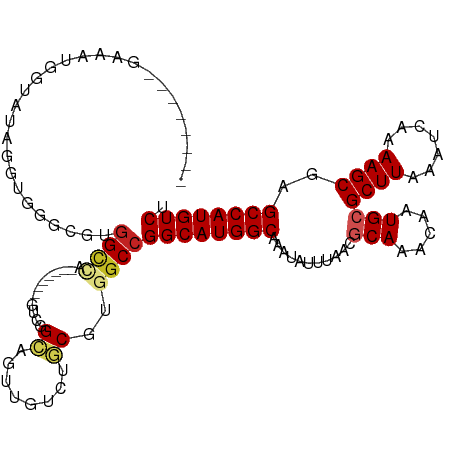
| Location | 26,199,145 – 26,199,258 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.63 |
| Shannon entropy | 0.53462 |
| G+C content | 0.49920 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.926426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26199145 113 - 27905053 AGACAUGGCUCGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGCCACGUUACCAUCCACCCCC------UCUGGCCAAACCCACCUAGACCAUUUCGCACAGCC .((.((((.((((.....((((((.((((.....))))))))))....))......(((((.................------..)))))...........)))))).))........ ( -23.91, z-score = -0.74, R) >droSim1.chr3R 25860213 110 - 27517382 AGACAUGGCUCGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGCCACGUUACCAACCACCCCC------UCUGGCCAAAC---CCUAGACCAUUUCGCACAGCA .((.((((.((((.....((((((.((((.....))))))))))....))......(((((.................------..)))))....---....)))))).))........ ( -23.91, z-score = -0.47, R) >droSec1.super_4 5039695 113 - 6179234 AGACAUGGCUCGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGCCACGUUACCACCCACCCCC------UCUGGCGAAACCCACCUAGACCAUUUCGCACAGCC .(((.(((((.((.....((((((.((((.....)))))))))).........)).))))).))).............------.(((((((((.............)))))).))).. ( -26.76, z-score = -1.57, R) >droYak2.chr3R 27131247 119 - 28832112 AGACAUGGCUCGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGCCACGUUACCACCCUCCUCCCCCCUUUUUGGCCAAACCCACCUAUACCAAUGCGCACAGCC .(.((((((..((((.......))))(((.....)))...........)))))).)(((((.........................)))))..................((.....)). ( -24.01, z-score = -0.73, R) >droEre2.scaffold_4820 8668702 99 + 10470090 AGACAUGGCUCGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGCCACGUUACCAGCCCCUCC---------UGGCCAGACCCACCUCCACCAA----------- .(.((((((..((((.......))))(((.....)))...........)))))).)(((((.(............).---------))))).................----------- ( -24.20, z-score = -1.28, R) >droAna3.scaffold_13340 21231587 103 - 23697760 AGACAUGGCUUGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGCCACGCAGAGCAUAGCCAAGU-------CAACC-CGCCCCUCUAUCCAACCUU-------- .(((.(((((((((((((((((((.((((.....))))))))))....(((.....)))..)).)))))).))))).))-------)....-...................-------- ( -29.70, z-score = -2.62, R) >dp4.chr2 29484765 103 + 30794189 AGACAUGGCUUGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGCCACGCAGACAACUGCAGACG--------AGCCACGUUCCGCUUUCCACGUUC-------- .(((.((((((((((.......))))(((((((((((((.........(((.....))).)))))))))).)))....)--------))))).)))...............-------- ( -31.80, z-score = -2.16, R) >droPer1.super_7 335017 103 + 4445127 AGACAUGGCUUGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGCCACGCAGACAACUGCAGACG--------AGCCACGUUCCGCUUUCCACGUUC-------- .(((.((((((((((.......))))(((((((((((((.........(((.....))).)))))))))).)))....)--------))))).)))...............-------- ( -31.80, z-score = -2.16, R) >droWil1.scaffold_181130 9701006 104 - 16660200 AGACAUGGCUUGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGACACGCAGACAAUUGCAGACG-------CCGCCACGUUUUGCGCCACACGGCC-------- ((((.((((..((((.......))))(((((((((((((..........((.....))..)))))))))).))).....-------..)))).))))...(((....))).-------- ( -30.90, z-score = -0.84, R) >droVir3.scaffold_12855 3321505 89 - 10161210 AGACAUGGCUCGCUUUUGAUUUAAGCGCAUUGUUUGGCUUAAAUAUUUGCCAUGCCGGCCACGCAGACAAUUGCAGACA--------UGCCACGUAC---------------------- ..((.((((..((((.......))))((((((((((((..........))))(((.(....))))))))).))).....--------.)))).))..---------------------- ( -23.10, z-score = -0.49, R) >droMoj3.scaffold_6540 24280144 89 + 34148556 AGACAUGGCUUGCUUUUGAUUUAAGCGCAUUGUUUGGGUUAAAUAUUUGCCAUGCCGGCCACGCAGACAAUUGCAGACA--------UGCCACGUAC---------------------- ..((.((((..((((.......))))((((((((((.((.........(((.....))).)).))))))).))).....--------.)))).))..---------------------- ( -24.40, z-score = -0.81, R) >consensus AGACAUGGCUCGCUUUUGAUUUAAGCGCAUUGUUUGCGUUAAAUAUUUGCCAUGCCGGCCACGCAGACAACCGCAGAC________UGGCCACGCCCACCUAUACCAGUUC________ .(.((((((.....((((((.(((((.....))))).)))))).....)))))).)(((.............................)))............................ (-16.90 = -17.27 + 0.36)

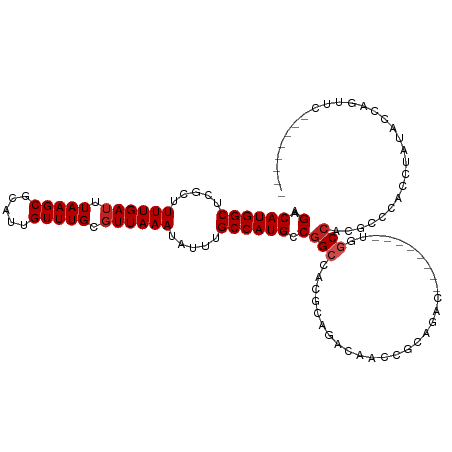
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:06 2011