| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,196,350 – 26,196,438 |
| Length | 88 |
| Max. P | 0.956827 |

| Location | 26,196,350 – 26,196,438 |
|---|---|
| Length | 88 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.13 |
| Shannon entropy | 0.51122 |
| G+C content | 0.56381 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -17.94 |
| Energy contribution | -15.89 |
| Covariance contribution | -2.05 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
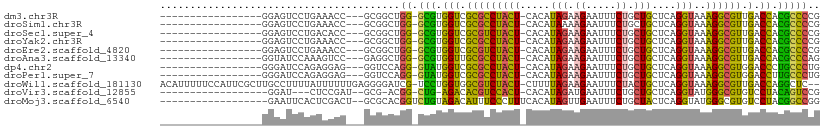
>dm3.chr3R 26196350 88 + 27905053 -----------------GGAGUCCUGAAACC---GCGGCUGG-GCGUGGUCGCGCCUACU-CACAUAGAAGAAUUUCUGCUGCUCAGGUAAAGGCGUUGACCACGCCCCG -----------------..((((.((....)---).))))((-(((((((((((((((((-....(((.((.....)).)))....)))..))))).))))))))))).. ( -37.00, z-score = -2.64, R) >droSim1.chr3R 25857479 88 + 27517382 -----------------GGAGUCCUGAAACC---GCGGCUGG-GCGUGGUCGCGCCUACU-CACAUAAAAGAAUUUCUGCUGCUCAGGUAAAGGCGUUGACCACGCCCCG -----------------..((((.((....)---).))))((-((((((((((((((..(-(........)).....((((.....)))).))))).))))))))))).. ( -35.40, z-score = -2.51, R) >droSec1.super_4 5036907 88 + 6179234 -----------------GGAGUCCUGACACC---GCGGCUGG-GCGUGGUCGCGUCUACU-CACAUAGAAGAAUUUCUGCUGCUCAGGUAAAGGCGUUGACCACGCCCCG -----------------..((((.((....)---).))))((-(((((((((((((((((-....(((.((.....)).)))....)))..))))).))))))))))).. ( -34.30, z-score = -1.72, R) >droYak2.chr3R 27128498 88 + 28832112 -----------------GGAGUCCUGAAACC---GCGGCUGG-GCGUGGUCGCGCCUACU-CACAUAGAAGAAUUUCUGCUGCUCAGGUAAAGGCGUUGACCACGCCCCG -----------------..((((.((....)---).))))((-(((((((((((((((((-....(((.((.....)).)))....)))..))))).))))))))))).. ( -37.00, z-score = -2.64, R) >droEre2.scaffold_4820 8666036 88 - 10470090 -----------------GGAGUCCUGAAACC---GCGGCUGG-GCGUGGUCGCGUCUACU-CACAUAGAAGAAUUUCUGCUGCUCAGGUAAAGGCGUUGACCACGCCCCG -----------------..((((.((....)---).))))((-(((((((((((((((((-....(((.((.....)).)))....)))..))))).))))))))))).. ( -34.30, z-score = -1.95, R) >droAna3.scaffold_13340 21228720 88 + 23697760 -----------------GGUAUCCAAAGUCC---GAGGCUGG-GCGUGGUUGCGCCUACU-CACAUAGAAGAAUUUCUGCUGCUCAGGUAAAGGCGUUGACCACGCCCAG -----------------..............---....((((-(((((((((((((((((-....(((.((.....)).)))....)))..))))).))))))))))))) ( -34.90, z-score = -2.66, R) >dp4.chr2 29481856 88 - 30794189 -----------------GGGAUCCAGAGGAG---GGUCCAGG-GUAUGGUCGCGCCUACU-CACAUAGAAGAAUUUCUGCUGCUCAGGUAAAGGCGUGGACCCUGCCCUG -----------------.((((((......)---)))))(((-(((.(((((((((((((-....(((.((.....)).)))....)))..)))))).)))).)))))). ( -37.90, z-score = -2.36, R) >droPer1.super_7 332108 88 - 4445127 -----------------GGGAUCCAGAGGAG---GGUCCAGG-GUAUGGUCGCGCCUACU-CACAUAGAAGAAUUUCUGCUGCUCAGGUAAAGGCGUGGACCUUGCCCUG -----------------.((((((......)---)))))(((-(((.(((((((((((((-....(((.((.....)).)))....)))..)))))).)))).)))))). ( -37.90, z-score = -2.62, R) >droWil1.scaffold_181130 9697521 106 + 16660200 ACAUUUUUCCAUUCGCUUGCCUUUUAUUUUUUGAGGGGAUCG-UCCUGGUGGCGUCUACU-CUUUUAGAAGAAUUUCUACUGCUCAGGUAAAGGCGUUGACCAGGCUC-- ...................(((((((.....)))))))...(-.(((((((((((((..(-(((....)))).....((((.....)))).))))))).)))))))..-- ( -26.20, z-score = -0.36, R) >droVir3.scaffold_12855 3317799 83 + 10161210 ------------------GGAU---CUCCGAU--GCG-ACGG-CUG-AGACACGUCCACU-CACAUAGAUGAAUUUCUGCUGCUCAGGUAUGGGCGUGUCCUACAGUCCG ------------------((..---..))...--...-.(((-(((-.((((((((((..-(.((((((......)))).)).....)..))))))))))...))).))) ( -26.60, z-score = -0.83, R) >droMoj3.scaffold_6540 24276434 90 - 34148556 ------------------GAAUUCACUCGACU--GCGCACGGUCUGUAGACAUUUCCCUUUCACAUAGUUGAAUUUCUGCUACUCAGGUAUGGGCGUGUCCUACGGCCGG ------------------..............--.....(((.((((((((((..(((.((((......))))..((((.....))))...))).)))).))))))))). ( -22.10, z-score = -0.21, R) >consensus _________________GGAAUCCUGAAACC___GCGGCUGG_GCGUGGUCGCGCCUACU_CACAUAGAAGAAUUUCUGCUGCUCAGGUAAAGGCGUUGACCACGCCCCG ........................................((.(((.(((.(((((((((.....(((.((.....)).)))....)))..)))))).).)).))))).. (-17.94 = -15.89 + -2.05)

| Location | 26,196,350 – 26,196,438 |
|---|---|
| Length | 88 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.13 |
| Shannon entropy | 0.51122 |
| G+C content | 0.56381 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -14.07 |
| Energy contribution | -14.06 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
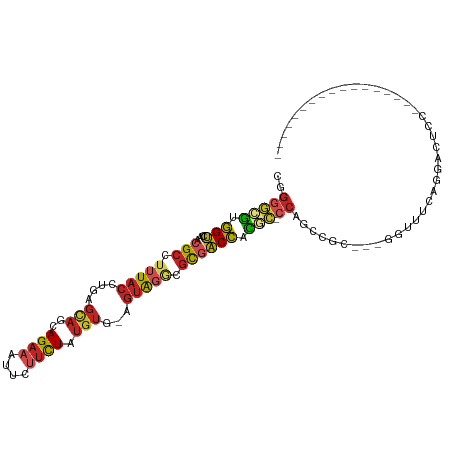
>dm3.chr3R 26196350 88 - 27905053 CGGGGCGUGGUCAACGCCUUUACCUGAGCAGCAGAAAUUCUUCUAUGUG-AGUAGGCGCGACCACGC-CCAGCCGC---GGUUUCAGGACUCC----------------- ..((((((((((..(((((....((..(((..((((....)))).))).-)).))))).))))))))-))((((..---.......)).))..----------------- ( -35.20, z-score = -2.09, R) >droSim1.chr3R 25857479 88 - 27517382 CGGGGCGUGGUCAACGCCUUUACCUGAGCAGCAGAAAUUCUUUUAUGUG-AGUAGGCGCGACCACGC-CCAGCCGC---GGUUUCAGGACUCC----------------- ..((((((((((..(((((((((.((((.((.(....).)))))).)))-)..))))).))))))))-))((((..---.......)).))..----------------- ( -34.40, z-score = -2.07, R) >droSec1.super_4 5036907 88 - 6179234 CGGGGCGUGGUCAACGCCUUUACCUGAGCAGCAGAAAUUCUUCUAUGUG-AGUAGACGCGACCACGC-CCAGCCGC---GGUGUCAGGACUCC----------------- ..((((((((((...((.(((((....(((..((((....)))).))).-.))))).))))))))))-))......---((.((....)).))----------------- ( -32.70, z-score = -1.39, R) >droYak2.chr3R 27128498 88 - 28832112 CGGGGCGUGGUCAACGCCUUUACCUGAGCAGCAGAAAUUCUUCUAUGUG-AGUAGGCGCGACCACGC-CCAGCCGC---GGUUUCAGGACUCC----------------- ..((((((((((..(((((....((..(((..((((....)))).))).-)).))))).))))))))-))((((..---.......)).))..----------------- ( -35.20, z-score = -2.09, R) >droEre2.scaffold_4820 8666036 88 + 10470090 CGGGGCGUGGUCAACGCCUUUACCUGAGCAGCAGAAAUUCUUCUAUGUG-AGUAGACGCGACCACGC-CCAGCCGC---GGUUUCAGGACUCC----------------- ..((((((((((...((.(((((....(((..((((....)))).))).-.))))).))))))))))-))((((..---.......)).))..----------------- ( -31.70, z-score = -1.50, R) >droAna3.scaffold_13340 21228720 88 - 23697760 CUGGGCGUGGUCAACGCCUUUACCUGAGCAGCAGAAAUUCUUCUAUGUG-AGUAGGCGCAACCACGC-CCAGCCUC---GGACUUUGGAUACC----------------- (((((((((((...(((((....((..(((..((((....)))).))).-)).)))))..)))))))-))))((..---.......)).....----------------- ( -34.50, z-score = -3.00, R) >dp4.chr2 29481856 88 + 30794189 CAGGGCAGGGUCCACGCCUUUACCUGAGCAGCAGAAAUUCUUCUAUGUG-AGUAGGCGCGACCAUAC-CCUGGACC---CUCCUCUGGAUCCC----------------- (((((.(((((((((((((....((..(((..((((....)))).))).-)).))))).........-..))))))---)).)))))......----------------- ( -31.70, z-score = -1.71, R) >droPer1.super_7 332108 88 + 4445127 CAGGGCAAGGUCCACGCCUUUACCUGAGCAGCAGAAAUUCUUCUAUGUG-AGUAGGCGCGACCAUAC-CCUGGACC---CUCCUCUGGAUCCC----------------- .((((...((((..(((((....((..(((..((((....)))).))).-)).))))).))))...)-)))(((.(---(......)).))).----------------- ( -28.70, z-score = -1.35, R) >droWil1.scaffold_181130 9697521 106 - 16660200 --GAGCCUGGUCAACGCCUUUACCUGAGCAGUAGAAAUUCUUCUAAAAG-AGUAGACGCCACCAGGA-CGAUCCCCUCAAAAAAUAAAAGGCAAGCGAAUGGAAAAAUGU --...((...((...((((((..........(((((....)))))...(-((..((((((....)).-)).))..)))........))))))....))..))........ ( -20.10, z-score = -0.14, R) >droVir3.scaffold_12855 3317799 83 - 10161210 CGGACUGUAGGACACGCCCAUACCUGAGCAGCAGAAAUUCAUCUAUGUG-AGUGGACGUGUCU-CAG-CCGU-CGC--AUCGGAG---AUCC------------------ (((.(((..(((((((.((((..(((.....)))....((((....)))-))))).)))))))-)))-))).-...--...((..---..))------------------ ( -24.70, z-score = -0.34, R) >droMoj3.scaffold_6540 24276434 90 + 34148556 CCGGCCGUAGGACACGCCCAUACCUGAGUAGCAGAAAUUCAACUAUGUGAAAGGGAAAUGUCUACAGACCGUGCGC--AGUCGAGUGAAUUC------------------ .((((.((((.(((..(((....(((.....)))...((((......)))).)))...))))))).).)))..(((--......))).....------------------ ( -19.80, z-score = 0.33, R) >consensus CGGGGCGUGGUCAACGCCUUUACCUGAGCAGCAGAAAUUCUUCUAUGUG_AGUAGGCGCGACCACGC_CCAGCCGC___GGUUUCAGGACUCC_________________ ..(((((.(((...(((.(((((....(((..((((....)))).)))...))))).)))))).))).))........................................ (-14.07 = -14.06 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:04 2011