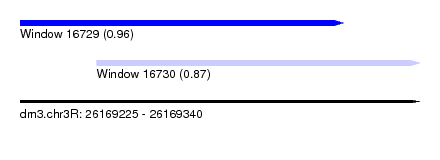
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,169,225 – 26,169,340 |
| Length | 115 |
| Max. P | 0.958558 |

| Location | 26,169,225 – 26,169,318 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 70.55 |
| Shannon entropy | 0.59067 |
| G+C content | 0.45889 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -13.11 |
| Energy contribution | -13.25 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
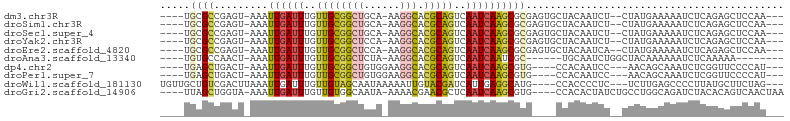
>dm3.chr3R 26169225 93 + 27905053 ----UGCGCCGAGU-AAAUUGAUUUGUUGCGGCUGCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUACAAUCU--CUAUGAAAAAUCUCAGAGCUCCAA--- ----.((((((.((-...((((((..((((((((...-..))).)))))..)))))))).)).))))......((--((..((....))..))))......--- ( -26.60, z-score = -0.76, R) >droSim1.chr3R 25831083 93 + 27517382 ----UGCGCCGAGU-AAAUUGAUUUGUUGCGGCUGCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUACAAUCU--CUAUGAAAAAUCUCAGAGCUCCAA--- ----.((((((.((-...((((((..((((((((...-..))).)))))..)))))))).)).))))......((--((..((....))..))))......--- ( -26.60, z-score = -0.76, R) >droSec1.super_4 5010502 93 + 6179234 ----UGCGCCGAGU-AAAUUGAUUUGUUGCGGCUGCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUACAAUCU--CUAUGAAAAAUCUCAGAGCUCCAA--- ----.((((((.((-...((((((..((((((((...-..))).)))))..)))))))).)).))))......((--((..((....))..))))......--- ( -26.60, z-score = -0.76, R) >droYak2.chr3R 27100924 93 + 28832112 ----UGCGCCGAGU-AAAUUGAUUUGUUGCGGCUCCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUACAAUCU--CUAUGAAAAAUCUCAGAGCUCCAA--- ----.((((((.((-...((((((..((((((((...-..))).)))))..)))))))).)).))))......((--((..((....))..))))......--- ( -26.00, z-score = -1.14, R) >droEre2.scaffold_4820 8639361 93 - 10470090 ----UGCGCCGAGU-AAAUUGAUUUGUUGCGGCUCCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUACAAUCA--CUAUGAAAAAUCUCAGAGCUCCAA--- ----.((((((.((-...((((((..((((((((...-..))).)))))..)))))))).)).))))........--((.(((......))).))......--- ( -23.90, z-score = -0.50, R) >droAna3.scaffold_13340 21202201 84 + 23697760 ----UGUGCCAACU-AAAUUGAUUUGUUGCGGCUCUA-AAGGCACGCAGUCAAUCAAUCGC------UGCAAUCUGGCUACAAAAAAUCUCAAAAA-------- ----(((((((...-..(((((((..((((((((...-..))).)))))..)))))))...------.......)))).)))..............-------- ( -20.16, z-score = -2.19, R) >dp4.chr2 29452335 89 - 30794189 ----UGAGCUGACU-AAAUUGAUUUGUUGCGGCUGUGGAAGGCACGCAGUCAAUCAAGCGUG----CCACAAUCC---AACAGCAAAUCUCGGUUCCCCAU--- ----.(((((((..-.....(((((((((.((...((...(((((((..........)))))----)).))..))---..)))))))))))))))).....--- ( -31.11, z-score = -2.74, R) >droPer1.super_7 302490 89 - 4445127 ----UGAGCUGACU-AAAUUGAUUUGUUGCGGCUGUGGAAGGCACGCAGUCAAUCAAGCGUG----CCACAAUCC---AACAGCAAAUCUCGGUUCCCCAU--- ----.(((((((..-.....(((((((((.((...((...(((((((..........)))))----)).))..))---..)))))))))))))))).....--- ( -31.11, z-score = -2.74, R) >droWil1.scaffold_181130 9666370 94 + 16660200 UGUUGCUGUCGACUUAAAUUGAUUUGUUGUAGCAAUAAAAAUUGUACGAUCAUUGAGGCAUG----CCACCCCUC---UCUUGAGCCCCUUAUGCUUCUAG--- ((((((((.((((............)))))))))))).................((((((((----......(((---....))).....))))))))...--- ( -22.10, z-score = -2.68, R) >droGri2.scaffold_14906 5974744 94 + 14172833 ----UUAGCUGGUA-AAAUUGAUUUGUUGUGGCAAUA-AAAACGAACGCUCAAUCAAGCGUG----CCACACUAUCUGCCUGGCAGAUCUACACAGUCAACUAA ----...(((((((-.....(((((((((.(((((((-.......(((((......))))).----......))).)))))))))))))))).))))....... ( -23.64, z-score = -1.68, R) >consensus ____UGCGCCGAGU_AAAUUGAUUUGUUGCGGCUGCA_AAGGCACGCAGUCAAUCAAGCGCG____CUACAAUCU__CUAUGAAAAAUCUCAGAGCUCCAA___ .....((((.........((((((..((((((((......))).)))))..))))))))))........................................... (-13.11 = -13.25 + 0.14)
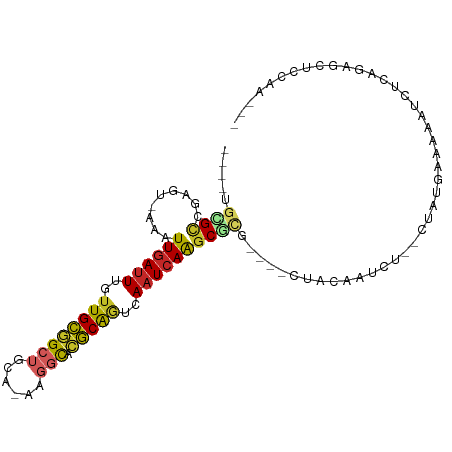
| Location | 26,169,247 – 26,169,340 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 61.42 |
| Shannon entropy | 0.77192 |
| G+C content | 0.51921 |
| Mean single sequence MFE | -20.02 |
| Consensus MFE | -8.52 |
| Energy contribution | -8.19 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26169247 93 + 27905053 UGCGGCUGCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUA-CAAUCUCUAUGAAAAAUCUCAGAGCUCCAACCCCAACUUCCCCCCCUAUUGC----- .((((((((.-.......))))))......((....))))..-....((((..((....))..))))............................----- ( -16.30, z-score = 0.45, R) >droSim1.chr3R 25831105 87 + 27517382 UGCGGCUGCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUA-CAAUCUCUAUGAAAAAUCUCAGAGCUCCAACUGCCCCUACUCCCUC----------- .((((((((.-.......)))))).....((((....)))).-....((((..((....))..))))........))............----------- ( -16.70, z-score = 0.49, R) >droSec1.super_4 5010524 87 + 6179234 UGCGGCUGCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUA-CAAUCUCUAUGAAAAAUCUCAGAGCUCCAACUGCCCCAACUCCCUC----------- .((((((((.-.......)))))).....((((....)))).-....((((..((....))..))))........))............----------- ( -16.70, z-score = 0.43, R) >droYak2.chr3R 27100946 98 + 28832112 UGCGGCUCCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUA-CAAUCUCUAUGAAAAAUCUCAGAGCUCCAACUUCCCUUCCCCAACUCUCCCGAUUGC (((((((...-..))).))))..(((((.((((....)))).-....((((..((....))..))))...........................))))). ( -18.50, z-score = -0.26, R) >droEre2.scaffold_4820 8639383 93 - 10470090 UGCGGCUCCA-AAGGCACGCAGUCAAUCAAGCGCGAGUGCUA-CAAUCACUAUGAAAAAUCUCAGAGCUCCAACUUCCCCAACUCCCCCCAGAGC----- ((.(((((..-..(((((.(.((.......))..).))))).-.........(((......)))))))).))..........(((......))).----- ( -15.50, z-score = 0.40, R) >dp4.chr2 29452357 78 - 30794189 UGCGGCUGUGGAAGGCACGCAGUCAAUCAAGCG----UGCCA-CAAUC-CAACAGCAAAUCUCGGUUCCCCAUCUAAAGCGGUG---------------- .((.((((((((.(((((((..........)))----)))).-...))-).))))).......((....)).......))....---------------- ( -26.40, z-score = -2.47, R) >droPer1.super_7 302512 78 - 4445127 UGCGGCUGUGGAAGGCACGCAGUCAAUCAAGCG----UGCCA-CAAUC-CAACAGCAAAUCUCGGUUCCCCAUCUAAAGCCGUG---------------- .((((((.((((.(((((((..........)))----)))).-.....-..............((....)).)))).)))))).---------------- ( -28.30, z-score = -3.63, R) >droWil1.scaffold_181130 9666397 91 + 16660200 UGUAGCAAUAAAAAUUGUACGAUCAUUGAGGCA----UGCCACCCCUCUCUUGAGCCCCUUAUGCUUCUAGUUCACUUAUCGAUCAGACUCGAUC----- .(((.((((....)))))))((.....((((((----((......(((....))).....))))))))....))....(((((......))))).----- ( -17.60, z-score = -1.08, R) >droVir3.scaffold_12855 3286484 86 + 10161210 UGUGGCAAUA-AAAGCGAACGUUCAAUCAAGCGU--UCGCCAGCAAUCUGCCUGGCAGAUCU-ACAGCUAACUCUCCCGCUAACUCUUUC---------- .((((.....-..((((((((((......)))))--))((((((.....).)))))......-...))).......))))..........---------- ( -24.14, z-score = -2.79, R) >consensus UGCGGCUGCA_AAGGCACGCAGUCAAUCAAGCGCGAGUGCUA_CAAUCUCUAUGAAAAAUCUCAGAGCUCCAACUCCCCCUACUCCCCC___________ (((((((......))).))))........((((....))))........................................................... ( -8.52 = -8.19 + -0.33)

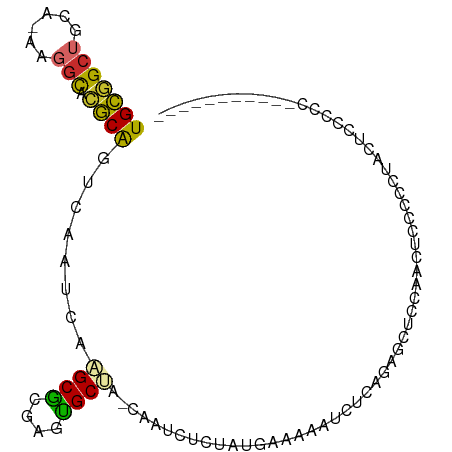
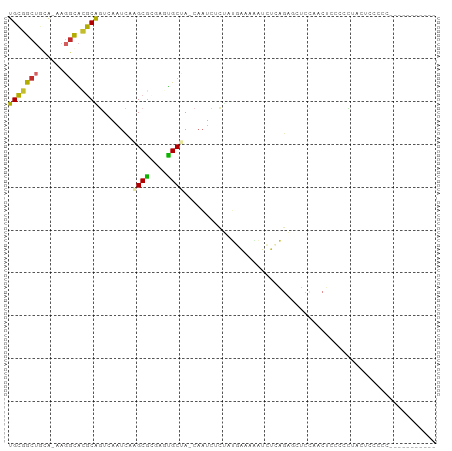
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:57:01 2011