
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,094,991 – 26,095,084 |
| Length | 93 |
| Max. P | 0.933257 |

| Location | 26,094,991 – 26,095,084 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Shannon entropy | 0.40573 |
| G+C content | 0.55874 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -19.83 |
| Energy contribution | -18.53 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
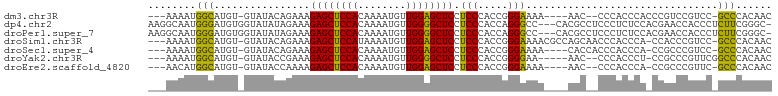
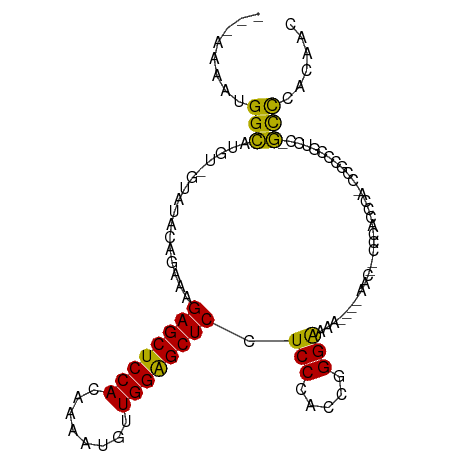
>dm3.chr3R 26094991 93 - 27905053 ---AAAAUGGCAUGU-GUAUACAGAAAGAGCUCCACAAAAUGUUGGAGCUCCUCCCACCGGGAAAA----AAC--CCCACCCACCCGUCCGUCC-GCCCACAAC ---...((((.(((.-((.........((((((((........))))))))........(((....----...--)))....)).)))))))..-......... ( -22.60, z-score = -2.23, R) >dp4.chr2 29381442 100 + 30794189 AAGGCAAUGGGAUGUGGUAUAUAGAAAGAGCUCCACAAAAUGUUGGGGCUCCUCCCACCAGGGCC---CACGCCUCCCUCUCCACGAACCACCCUCUUCGGGC- ..((....(((((((((..........((((((((........))))))))..(((....))).)---))))..))))...))........(((.....))).- ( -32.50, z-score = -0.31, R) >droPer1.super_7 232330 100 + 4445127 AAGGCAAUGGGAUGUGGUAUAUAGAAAGAGCUCCACAAAAUGUUGGGGCUCCUCCCACCAGGGCC---CACGCCUCCCUCUCCACGAACCACCCUCUUCGGGC- ..((....(((((((((..........((((((((........))))))))..(((....))).)---))))..))))...))........(((.....))).- ( -32.50, z-score = -0.31, R) >droSim1.chr3R 25762373 98 - 27517382 ---AAAAUGGCAUGU-GUAUACAGAAAGAGCUCCAUAAAAUGUUGGAGCUCCUCCCACCGGGAAAACGCCAGCAACCCACCCA-CCACCCGUCC-GCCCACAAC ---....((((....-...........((((((((........)))))))).((((...))))....))))............-..........-......... ( -24.80, z-score = -2.54, R) >droSec1.super_4 4945295 94 - 6179234 ---AAAAUGGCAUGU-GUAUACAGAAAGAGCUCCACAAAAUGUUGGAGCUCCUCCCACCGGGAAAA----CACCACCCACCCA-CCGCCCGUCC-GCCCACAAC ---....(((...((-((.........((((((((........)))))))).((((...))))..)----)))...)))....-..........-......... ( -23.20, z-score = -2.11, R) >droYak2.chr3R 27030788 92 - 28832112 ---AAAAUGGCAUGU-GUAUACCGAAAGAGCUCCACAAAAUGUUGGGGCUCCUCCCACCGGGGAA-----AAC--CCCACCCU-CCGCCCGUUCGGCCCACAAC ---.........(((-(....(((((.((((((((........))))))))........((((..-----..)--))).....-.......)))))..)))).. ( -28.80, z-score = -1.27, R) >droEre2.scaffold_4820 8571459 92 + 10470090 ---AACAUGGCAUGU-GUAUACCAAAAGAGCUCCACAAAAUGUUGGAGCUCCUCCCACCGGGAAAA----AAC--CCCACCCA-CCGCCCGUUC-GCCCACAAC ---.....(((..((-(..........((((((((........))))))))........(((....----...--)))...))-).))).....-......... ( -23.50, z-score = -2.54, R) >consensus ___AAAAUGGCAUGU_GUAUACAGAAAGAGCUCCACAAAAUGUUGGAGCUCCUCCCACCGGGAAAA____AAC__CCCACCCA_CCGCCCGUCC_GCCCACAAC ........(((................((((((((........)))))))).(((.....)))................................)))...... (-19.83 = -18.53 + -1.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:56 2011