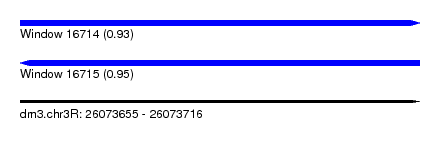
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,073,655 – 26,073,716 |
| Length | 61 |
| Max. P | 0.945753 |

| Location | 26,073,655 – 26,073,716 |
|---|---|
| Length | 61 |
| Sequences | 11 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 71.20 |
| Shannon entropy | 0.61911 |
| G+C content | 0.44138 |
| Mean single sequence MFE | -10.75 |
| Consensus MFE | -8.48 |
| Energy contribution | -8.57 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.55 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
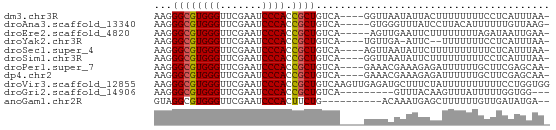
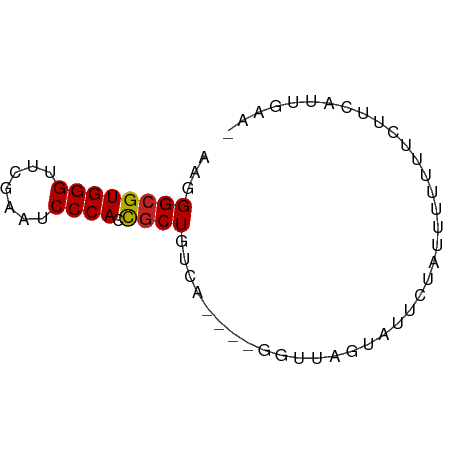
>dm3.chr3R 26073655 61 + 27905053 -UUAAAUGAGGAAAAAAAAGUAAUAUUAACC----UGACAGCGGUGGGAUUCGAACCCACGCCCUU -......((((........((..........----..)).(((.((((.......))))))))))) ( -10.30, z-score = -0.38, R) >droAna3.scaffold_13340 21114509 60 + 23697760 -CUUAACAAAAAAUGUAAGGAUAAACCCAC-----UGACAGCGGUGGGAUUCGAACCCACGCCCUU -...............((((.....(((((-----((....)))))))...((......)).)))) ( -13.10, z-score = -1.29, R) >droEre2.scaffold_4820 8547562 60 - 10470090 -UUCAAUUAUCUAAAAAAAAGAAUUCAACU-----UGACAGCGGUGGGAUUCGAACCCACGCCCUU -.............................-----.....(((.((((.......))))))).... ( -9.10, z-score = -0.35, R) >droYak2.chr3R 27007523 58 + 28832112 -UUAAAUGAGGAAAAAAA--GAAU-UCAACA----UGACAGCGGUGGGAUUCGAACCCACGCCCUU -......((((.......--....-((....----.))..(((.((((.......))))))))))) ( -11.10, z-score = -0.80, R) >droSec1.super_4 4924047 61 + 6179234 -UUAAAUGAGAAAAAAAAAAGAAUAUUAACU----UGACAGCGGUGGGAUUCGAACCCACGCCCUU -..............................----.....(((.((((.......))))))).... ( -9.10, z-score = -0.80, R) >droSim1.chr3R 25739734 61 + 27517382 -UUAAAUGAGGAAAAAAAAAGAAUAUUAACC----UGACAGCGGUGGGAUUCGAACCCACGCCCUU -......((((........((.........)----)....(((.((((.......))))))))))) ( -10.00, z-score = -0.49, R) >droPer1.super_7 210799 61 - 4445127 -UUGCUCGAAGCAAAAAAUCUCUUUCGUUUC----UGACAGCGGUGGGAUUCGAACCCACGCCCUU -.((((.(((((.(((......))).)))))----....))))(((((.......)))))...... ( -13.40, z-score = -0.28, R) >dp4.chr2 29359969 61 - 30794189 -UUGCUCGAAGCAAAAAAUCUCUUUCGUUUC----UGACAGCGGUGGGAUUCGAACCCACGCCCUU -.((((.(((((.(((......))).)))))----....))))(((((.......)))))...... ( -13.40, z-score = -0.28, R) >droVir3.scaffold_12855 3168654 66 + 10161210 CCACCAGGAAAAAAAAAAUAGAAAGCAUCUCAACUUGACAGCGGUGGGAUUCGAACCCACGCCCUU .....(((................((.((.......))..)).(((((.......)))))..))). ( -10.80, z-score = 0.05, R) >droGri2.scaffold_14906 5858775 54 + 14172833 ---CCACCAAAAAUAAACUUGUAAAC---------UGACAGCGGUGGGAUUCGAACCCACGCCCUU ---.......................---------.....(((.((((.......))))))).... ( -9.10, z-score = -0.34, R) >anoGam1.chr2R 37002181 54 - 62725911 --UCAUAUCAACAAAAAAGCUCAUUUGU----------CAGAAGUGGGAUUCGAACCCACGCCUAC --.....((.(((((........)))))----------..)).(((((.......)))))...... ( -8.90, z-score = -1.10, R) >consensus _UUAAAUGAAAAAAAAAAAAGAAUACGAACC____UGACAGCGGUGGGAUUCGAACCCACGCCCUU ........................................((.(((((.......))))))).... ( -8.48 = -8.57 + 0.09)

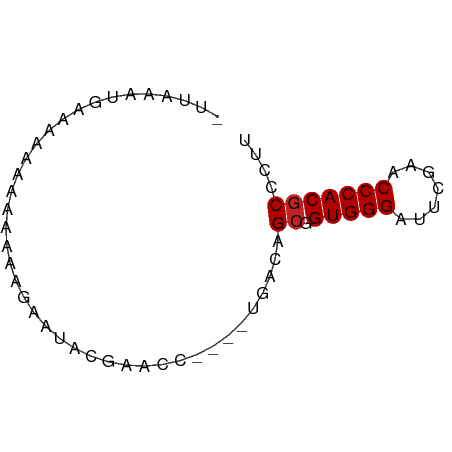
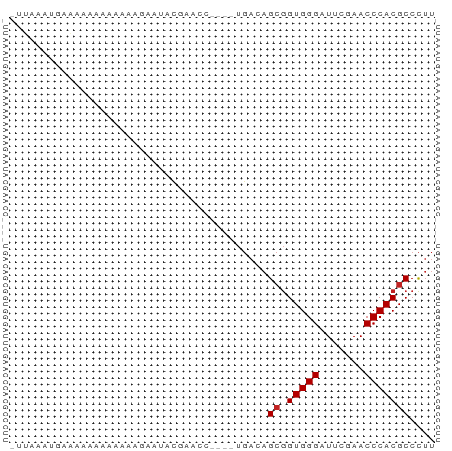
| Location | 26,073,655 – 26,073,716 |
|---|---|
| Length | 61 |
| Sequences | 11 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 71.20 |
| Shannon entropy | 0.61911 |
| G+C content | 0.44138 |
| Mean single sequence MFE | -13.52 |
| Consensus MFE | -10.58 |
| Energy contribution | -10.59 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26073655 61 - 27905053 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA----GGUUAAUAUUACUUUUUUUUCCUCAUUUAA- ...((((((((.......)))).))))...(----((..((..........))..))).......- ( -11.70, z-score = -0.52, R) >droAna3.scaffold_13340 21114509 60 - 23697760 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA-----GUGGGUUUAUCCUUACAUUUUUUGUUAAG- ...((((((((.......)))).))))....-----(((((......))))).............- ( -14.50, z-score = -0.76, R) >droEre2.scaffold_4820 8547562 60 + 10470090 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA-----AGUUGAAUUCUUUUUUUUAGAUAAUUGAA- ...((((((((.......)))).)))).(((-----(.(((((........)))))....)))).- ( -13.90, z-score = -0.89, R) >droYak2.chr3R 27007523 58 - 28832112 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA----UGUUGA-AUUC--UUUUUUUCCUCAUUUAA- ...((((((((.......)))).))))...(----((..((-(...--.....)))..)))....- ( -12.90, z-score = -1.03, R) >droSec1.super_4 4924047 61 - 6179234 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA----AGUUAAUAUUCUUUUUUUUUUCUCAUUUAA- ...((((((((.......)))).))))....----..............................- ( -11.20, z-score = -1.21, R) >droSim1.chr3R 25739734 61 - 27517382 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA----GGUUAAUAUUCUUUUUUUUUCCUCAUUUAA- ...((((((((.......)))).))))...(----((..((..........))..))).......- ( -11.70, z-score = -0.65, R) >droPer1.super_7 210799 61 + 4445127 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA----GAAACGAAAGAGAUUUUUUGCUUCGAGCAA- ...((((((((.......)))).))))((..----(((.((((((....)))))).)))..))..- ( -16.80, z-score = -0.56, R) >dp4.chr2 29359969 61 + 30794189 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA----GAAACGAAAGAGAUUUUUUGCUUCGAGCAA- ...((((((((.......)))).))))((..----(((.((((((....)))))).)))..))..- ( -16.80, z-score = -0.56, R) >droVir3.scaffold_12855 3168654 66 - 10161210 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCAAGUUGAGAUGCUUUCUAUUUUUUUUUUCCUGGUGG .((((((((((.......)))).)))....((((......))))..............)))..... ( -13.20, z-score = 0.69, R) >droGri2.scaffold_14906 5858775 54 - 14172833 AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA---------GUUUACAAGUUUAUUUUUGGUGG--- ...((((((((.......)))).))))....---------.....((((......))))....--- ( -12.30, z-score = -0.36, R) >anoGam1.chr2R 37002181 54 + 62725911 GUAGGCGUGGGUUCGAAUCCCACUUCUG----------ACAAAUGAGCUUUUUUGUUGAUAUGA-- .((((.(((((.......))))).))))----------.((.((.(((......))).)).)).-- ( -13.70, z-score = -1.50, R) >consensus AAGGGCGUGGGUUCGAAUCCCACCGCUGUCA____GGUUAGUAUUCUAUUUUUUUCUUCAUUGAA_ ...((((((((.......)))).))))....................................... (-10.58 = -10.59 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:49 2011