| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,072,127 – 26,072,243 |
| Length | 116 |
| Max. P | 0.753736 |

| Location | 26,072,127 – 26,072,243 |
|---|---|
| Length | 116 |
| Sequences | 9 |
| Columns | 126 |
| Reading direction | reverse |
| Mean pairwise identity | 64.38 |
| Shannon entropy | 0.72495 |
| G+C content | 0.48613 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -11.78 |
| Energy contribution | -11.95 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.753736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
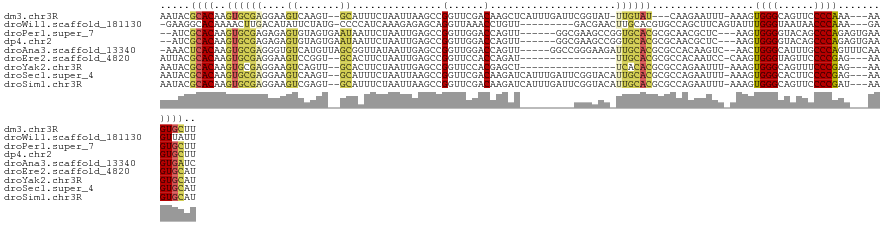
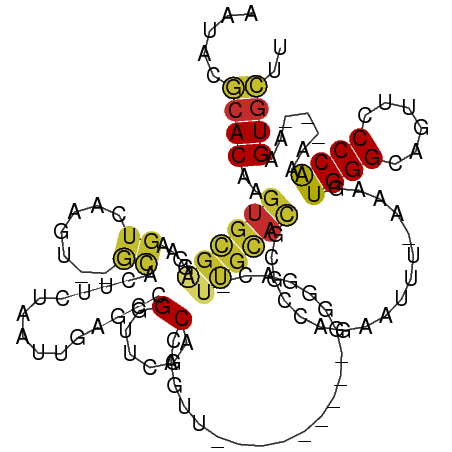
>dm3.chr3R 26072127 116 - 27905053 AAUACGCACAAGUGCGAGGAAGUCAAGU--GCAUUUCUAAUUAAGCCGGUUCGACAAGCUCAUUUGAUUCGGUAU-UUGUAU---CAAGAAUUU-AAAGUGGGCAGUUCCCCAAA---AAGUGCUU .....(((((((((((..(....)...)--))))))...........((...(((..(((((((((((((((((.-...)))---)..))))).-.)))))))).)))..))...---..)))).. ( -25.60, z-score = -0.49, R) >droWil1.scaffold_181130 9546827 112 - 16660200 -GAAGGCACAAAACUUGACAUAUUCUAUG-CCCCAUCAAAGAGAGCAGGUUAAACCUGUU---------GACGAACUUGCACGUGCCAGCUUCAGUAUUUGGGUAAUAACCCAAA---GAGUUAUU -...(((((.............((((.((-......)).))))((((((.....))))))---------.............)))))(((((.....(((((((....)))))))---)))))... ( -28.60, z-score = -1.96, R) >droPer1.super_7 209273 115 + 4445127 --AUCGCACAAGUGCGAGAGAGUGUAGUGAAUAAUUCUAAUUGAGCCGGUUGGACCAGUU------GGCGAAGCCGGUGCACGCGCAACGCUC---AAGUGGGGUACAGCCCAGAGUGAAGUGCUU --...((((..(((((..(((((..........)))))...((.(((((((...((....------))...))))))).)))))))..(((((---...((((......)))))))))..)))).. ( -41.80, z-score = -2.04, R) >dp4.chr2 29358439 115 + 30794189 --AUCGCACAAGUGCGAGAGAGUGUAGUGAAUAAUUCUAAUUGAGCCGGUUGGACCAGUU------GGCGAAGCCGGUGCACGCGCAACGCUC---AAGUGGGGUACAGCCCAGAGUGAAGUGCUU --...((((..(((((..(((((..........)))))...((.(((((((...((....------))...))))))).)))))))..(((((---...((((......)))))))))..)))).. ( -41.80, z-score = -2.04, R) >droAna3.scaffold_13340 21112941 118 - 23697760 -AAACUCACAAGUGCGAGGGUGUCAUGUUAGCGGUUAUAAUUGAGCCGGUUGGACCAGUU-----GGCCGGGAAGAUUGCACGCGCCACAAGUC--AACUGGGCAUUUGCCCAGUUUCAAGUGAUC -....((((..(((.(.(.((((.((.((..((((..((((((..((....))..)))))-----)))))..)).)).)))).).)))).....--((((((((....))))))))....)))).. ( -41.30, z-score = -1.75, R) >droEre2.scaffold_4820 8546027 104 + 10470090 AUUACGCACAAGUGCGAGGAAGUCCGGU--GCACUUCUAAUUGAGCCGGUUCCACCAGAU----------------UUGCACGCGCCACAAUCC-CAAGUGGGUAGUUCCCCGAG---AAGUGCAU ....((((....)))).((....)).((--((((((((....((((.((.....))....----------------......((.((((.....-...)))))).))))....))---)))))))) ( -33.30, z-score = -1.87, R) >droYak2.chr3R 27005975 104 - 28832112 AAUACGCACAAGUGCGAGGAAGUCAGUU--GCACUUCUAAUUGAGCCGGUUCCACGAGCU----------------UCACACGCGCCAGAAUUU-AAAGUGGGCAGUUUCCCGAG---AAGUGCAU .....((((..(((((.(((((((....--).))))))....((((((......)).)))----------------)....)))))........-....((((......))))..---..)))).. ( -28.80, z-score = -0.75, R) >droSec1.super_4 4922518 120 - 6179234 AAUACGCACAAGUGCGAGGAAGUCAAGU--GCAUUUCUAAUUAAGCCGGUUCGACAAGAUCAUUUGAUUCGGUACAUUGCACGCGCCAGAAUUU-AAAGUGGGCACUUCCCCGAG---AAGUGCAU .....((((.....((.((((((...((--(((...........((((..((((.........))))..))))....)))))((.(((......-....))))))))))).))..---..)))).. ( -32.26, z-score = -1.18, R) >droSim1.chr3R 25738192 120 - 27517382 AAUACGCACAAGUGCGAGGAAGUCGAGU--GCAUUUCUAAUUAAGCCGGUUCGACAAGAUCAUUUGAUUCGGUACAUUGCACGCGCCAGAAUUU-AAAGUGGGCAGUUCCCCGAU---AAGUGCAU .....((((..((((((((((((.....--..))))))......((((..((((.........))))..))))...))))))((.(((......-....)))))...........---..)))).. ( -28.90, z-score = -0.03, R) >consensus AAUACGCACAAGUGCGAGGAAGUCAAGU__GCACUUCUAAUUGAGCCGGUUCGACCAGUU_________GGGGAC_UUGCACGCGCCAGAAUUU_AAAGUGGGCAGUUCCCCAAA___AAGUGCUU .....((((..((((((...........................................................)))))).................((((......)))).......)))).. (-11.78 = -11.95 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:47 2011