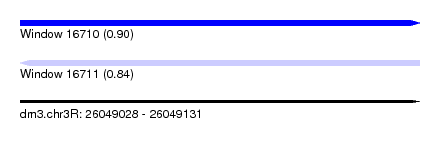
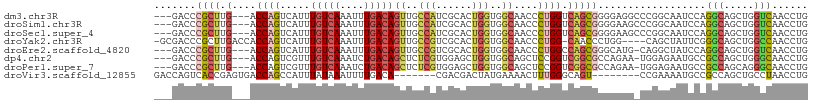
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,049,028 – 26,049,131 |
| Length | 103 |
| Max. P | 0.904220 |

| Location | 26,049,028 – 26,049,131 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.54 |
| Shannon entropy | 0.53624 |
| G+C content | 0.58280 |
| Mean single sequence MFE | -43.62 |
| Consensus MFE | -14.09 |
| Energy contribution | -14.45 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
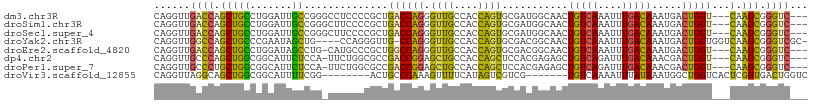
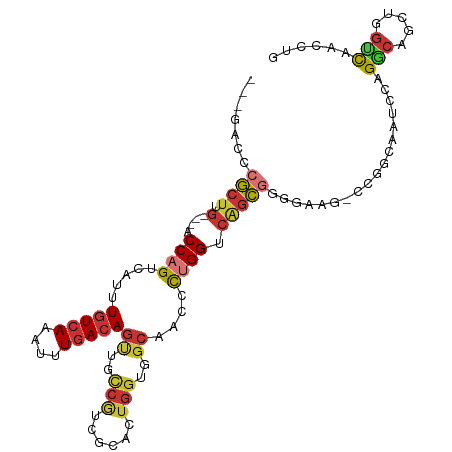
>dm3.chr3R 26049028 103 + 27905053 CAGGUUGACCAGCUGCCUGGAUUGCCGGGCCUCCCCGCUGACCAGGGUUGCCACCAGUGCGAUGGCAACUGUCAAAUUUGACAAAUGACUGGU---CAAGCGGGUC--- ..((....))....((((((....))))))...((((((((((((((((((((.(.....).))))))))((((....))))......)))))---).))))))..--- ( -49.10, z-score = -3.63, R) >droSim1.chr3R 25716394 103 + 27517382 CAGGUUGACCAGCUGCCUGGAUUGCCGGGCUUCCCCGCUGACCAGGGUUGCCACCAGUGCGAUGGCAACUGUCAAAUUUGACAAAUGACUGGU---CAAGCGGGUC--- ..((....))....((((((....))))))...((((((((((((((((((((.(.....).))))))))((((....))))......)))))---).))))))..--- ( -49.50, z-score = -3.92, R) >droSec1.super_4 4900485 103 + 6179234 CAGGUUGACCAGCUGCCUGGAUUGCCGGGCUUCCCCGCUGACCAGGGUUGCCACCAGUGCGAUGGCAACUGUCAAAUUUGACAAAUGACUGGU---CAAGCGGGUC--- ..((....))....((((((....))))))...((((((((((((((((((((.(.....).))))))))((((....))))......)))))---).))))))..--- ( -49.50, z-score = -3.92, R) >droYak2.chr3R 26982666 103 + 28832112 CAGGUUGGCCAGCUGCCCGAAUAGCUG----CCAGGGUUG-CCAGGGUUGCCACCAGUGCGACGGCAACUGUCAAAUUUGACAAAUGACUGGUGGUCAAGCGGGUCGC- ..((....)).((.(((((....((..----(((((((((-((...((((((....).))))))))))))((((....))))......))))..))....))))).))- ( -43.70, z-score = -1.74, R) >droEre2.scaffold_4820 8524190 102 - 10470090 CAGGUUGACCAGCUGCCUGGAUAGCCUG-CAUGCCCGCUGGCCAGGGUUGCCACCAGUGCGACGGCAACUGUCAAAUUUGACAAAUGACUGGU---CAAGCGGGUC--- (((((((.((((....)))).)))))))-...((((((((((((((((((((...........)))))))((((....))))......)))))---).))))))).--- ( -49.90, z-score = -3.91, R) >dp4.chr2 29336077 102 - 30794189 CAGGUUGCCCAGCUGGCGGCAUUCUCCA-UUCUGGCGCCGACCGGAGCUGCCACCAGCUCCACGAGAGCUGUCAGAUUUGACAAACGACUGGU---CAAGCGGGUC--- ......((((.(((((((.((.......-...)).))))(((((((((((....)))))))........((((......)))).......)))---).))))))).--- ( -40.70, z-score = -1.39, R) >droPer1.super_7 186880 102 - 4445127 CAGGUUGCCCUGCUGGCGGCAUUCUCCA-UUCUGGCGCCGACCGGAGCUGCCACCAGCUCCACGAGAGCUGUCAGAUUUGACAAACGACUGGU---CAAGCGGGUC--- ........((((((((((.((.......-...)).))))(((((((((((....)))))))........((((......)))).......)))---).))))))..--- ( -40.90, z-score = -1.36, R) >droVir3.scaffold_12855 7023305 94 - 10161210 CAGGUUAGGCAGCUGGCGGCAUUUUCGG--------ACUGCCCAAAGUUUUCAUAGUCGUCG-------UGUCAAAAUUUAUAAAUGGCUGGUCACUCGGUGACUGGUC .......((((..(((((((......((--------(((......))))).....)))))))-------)))).............(((..(((((...)))))..))) ( -25.70, z-score = -0.93, R) >consensus CAGGUUGACCAGCUGCCUGGAUUGCCGG_CUUCCCCGCUGACCAGGGUUGCCACCAGUGCGACGGCAACUGUCAAAUUUGACAAAUGACUGGU___CAAGCGGGUC___ ..((..(.((((....)))).)..)).............((((...(((...((((((.....(....)(((((....)))))....)))))).....))).))))... (-14.09 = -14.45 + 0.36)
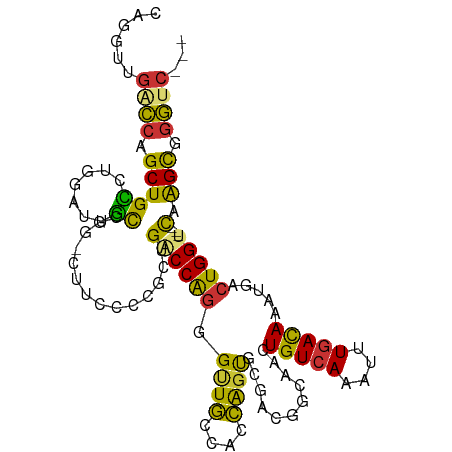
| Location | 26,049,028 – 26,049,131 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.54 |
| Shannon entropy | 0.53624 |
| G+C content | 0.58280 |
| Mean single sequence MFE | -40.61 |
| Consensus MFE | -13.70 |
| Energy contribution | -14.11 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.837923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26049028 103 - 27905053 ---GACCCGCUUG---ACCAGUCAUUUGUCAAAUUUGACAGUUGCCAUCGCACUGGUGGCAACCCUGGUCAGCGGGGAGGCCCGGCAAUCCAGGCAGCUGGUCAACCUG ---..((((((.(---(((((.....(((((....)))))(((((((((.....))))))))).)))))))))))).(((.(((((..........)))))....))). ( -48.30, z-score = -3.65, R) >droSim1.chr3R 25716394 103 - 27517382 ---GACCCGCUUG---ACCAGUCAUUUGUCAAAUUUGACAGUUGCCAUCGCACUGGUGGCAACCCUGGUCAGCGGGGAAGCCCGGCAAUCCAGGCAGCUGGUCAACCUG ---..((((((.(---(((((.....(((((....)))))(((((((((.....))))))))).))))))))))))...(((.((....)).)))....((....)).. ( -47.60, z-score = -3.92, R) >droSec1.super_4 4900485 103 - 6179234 ---GACCCGCUUG---ACCAGUCAUUUGUCAAAUUUGACAGUUGCCAUCGCACUGGUGGCAACCCUGGUCAGCGGGGAAGCCCGGCAAUCCAGGCAGCUGGUCAACCUG ---..((((((.(---(((((.....(((((....)))))(((((((((.....))))))))).))))))))))))...(((.((....)).)))....((....)).. ( -47.60, z-score = -3.92, R) >droYak2.chr3R 26982666 103 - 28832112 -GCGACCCGCUUGACCACCAGUCAUUUGUCAAAUUUGACAGUUGCCGUCGCACUGGUGGCAACCCUGG-CAACCCUGG----CAGCUAUUCGGGCAGCUGGCCAACCUG -((..((((.(((.((((((((...((((((....))))))..((....)))))))))))))..(((.-((....)).----))).....))))..))........... ( -34.00, z-score = -0.37, R) >droEre2.scaffold_4820 8524190 102 + 10470090 ---GACCCGCUUG---ACCAGUCAUUUGUCAAAUUUGACAGUUGCCGUCGCACUGGUGGCAACCCUGGCCAGCGGGCAUG-CAGGCUAUCCAGGCAGCUGGUCAACCUG ---(.((((((.(---.((((.....(((((....)))))((((((..(.....)..)))))).))))).)))))))...-((((....((((....))))....)))) ( -42.30, z-score = -2.16, R) >dp4.chr2 29336077 102 + 30794189 ---GACCCGCUUG---ACCAGUCGUUUGUCAAAUCUGACAGCUCUCGUGGAGCUGGUGGCAGCUCCGGUCGGCGCCAGAA-UGGAGAAUGCCGCCAGCUGGGCAACCUG ---...((((..(---(....))..((((((....)))))).....))))(((((((((((.(((((.((.......)).-)))))..)))))))))))((....)).. ( -43.80, z-score = -1.93, R) >droPer1.super_7 186880 102 + 4445127 ---GACCCGCUUG---ACCAGUCGUUUGUCAAAUCUGACAGCUCUCGUGGAGCUGGUGGCAGCUCCGGUCGGCGCCAGAA-UGGAGAAUGCCGCCAGCAGGGCAACCUG ---...((((..(---(....))..((((((....)))))).....)))).((((((((((.(((((.((.......)).-)))))..))))))))))(((....))). ( -44.40, z-score = -2.10, R) >droVir3.scaffold_12855 7023305 94 + 10161210 GACCAGUCACCGAGUGACCAGCCAUUUAUAAAUUUUGACA-------CGACGACUAUGAAAACUUUGGGCAGU--------CCGAAAAUGCCGCCAGCUGCCUAACCUG .....(((((...)))))(((...((((((....(((...-------...))).))))))....(((((((((--------.((.......))...))))))))).))) ( -16.90, z-score = -0.51, R) >consensus ___GACCCGCUUG___ACCAGUCAUUUGUCAAAUUUGACAGUUGCCGUCGCACUGGUGGCAACCCUGGUCAGCGGGGAAG_CCGGCAAUCCAGGCAGCUGGUCAACCUG .......((((......((((.....(((((....)))))((..(((......)))..))....))))..))))..................(((.....)))...... (-13.70 = -14.11 + 0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:46 2011