| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,205,552 – 9,205,644 |
| Length | 92 |
| Max. P | 0.719407 |

| Location | 9,205,552 – 9,205,644 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 71.96 |
| Shannon entropy | 0.54646 |
| G+C content | 0.38459 |
| Mean single sequence MFE | -16.27 |
| Consensus MFE | -8.88 |
| Energy contribution | -8.71 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
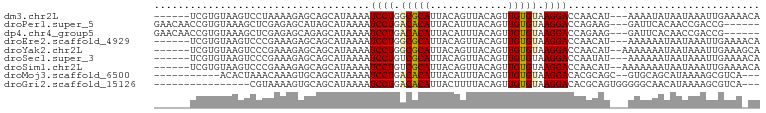
>dm3.chr2L 9205552 92 - 23011544 ------UCGUGUAAGUCCUAAAAGAGCAGCAUAAAAUCCUGGCGCAUUACAGUUACAGUUGUGUAAGGACCAACAU---AAAAUAUAAUAAAUUGAAAACA ------..((((..(((((.(....(((((........(((........))).....))))).).)))))..))))---...................... ( -14.52, z-score = -0.66, R) >droPer1.super_5 5932476 92 - 6813705 GAACAACCGUGUAAAGCUCGAGAGCAUAGCAUAAAAUCCUGACACAUUACAUUUACAGUUGUGUAAGGACCAGAAG---GAUUCACAACCGACCG------ ......((((((...(((....)))...))))....((((.(((((.............))))).))))......)---)...............------ ( -16.42, z-score = -0.95, R) >dp4.chr4_group5 1523756 92 - 2436548 GAACAACCGUGUAAAGCUCGAGAGCAGAGCAUAAAAUCCUGACACAUUACAUUUACAGUUGUGUAAGGACCAGAAG---GAUUCACAACCGACCG------ ........(((....((((.......))))....((((((......((((((........))))))........))---))))))).........------ ( -18.24, z-score = -1.45, R) >droEre2.scaffold_4929 9814900 92 - 26641161 ------UCGUGUAAGUCCCGAAAGAGCAGCAUAAAAUCCUGGCGCAUUACAGUUACAGUUGUGUAAGGACCAACAU---AAAAAAUAAUAAAUUGAAAACA ------..((((..(((((....)....((((((....(((........)))......))))))..))))..))))---...................... ( -15.90, z-score = -1.26, R) >droYak2.chr2L 11876275 93 - 22324452 ------UCGUGUAAGUCCCGAAAGAGCAGCAUAAAAUCCUGGCGCAUUACAGUUACAGUUGUGUAAGGACCAACAU--AAAAAAAUAAUAAAUUGAAAGCA ------..((((..(((((....)....((((((....(((........)))......))))))..))))..))))--....................... ( -15.90, z-score = -1.03, R) >droSec1.super_3 4669866 92 - 7220098 ------UCGUGUAAGUCCCGAAAGAGCAGCAUAAAAUCCUGUCGCAUUACAGUUACAGUUGUGUAAGGACCAAUAU---AAAAAAUAAUAAAUUGAAAACA ------..((((..(((((....)....((((((....((((.((......)).))))))))))..))))..))))---...................... ( -15.30, z-score = -1.89, R) >droSim1.chr2L 8986765 93 - 22036055 ------UCGUGUAAGUCCCGAAAGAGCAGCAUAAAAUCCUGUCGCAUUACAGUUACAGUUGUGUAAGGACCAACAU--AAAAAAAUAAUAAAUUGAAAACA ------..((((..(((((....)....((((((....((((.((......)).))))))))))..))))..))))--....................... ( -17.20, z-score = -2.46, R) >droMoj3.scaffold_6500 12118565 85 - 32352404 -----------ACACUAAACAAAGUGCAGCAUAAAAUCCUGACACAUUACAUUUACAGUUGUGUAAGGACACGCAGC--GUGCAGCAUAAAAGCGUCA--- -----------.............((((((......((((.(((((.............))))).))))......))--.))))((......))....--- ( -16.82, z-score = -0.03, R) >droGri2.scaffold_15126 5387720 82 + 8399593 ----------------CGUAAAAGUGCAGCAUAAAAUCCUGACACAUUACUUUUACAGUUGUGUAAGGACACGCAGUGGGGGCAACAUAAAAGCGUCA--- ----------------.(((....))).........((((.(((((..(((.....)))))))).)))).((((..(...(....)...)..))))..--- ( -16.10, z-score = 0.17, R) >consensus ______UCGUGUAAGUCCCGAAAGAGCAGCAUAAAAUCCUGACGCAUUACAGUUACAGUUGUGUAAGGACCAACAU___AAAAAAUAAUAAAUUGAAA_CA ....................................((((.(((((.............))))).))))................................ ( -8.88 = -8.71 + -0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:44 2011