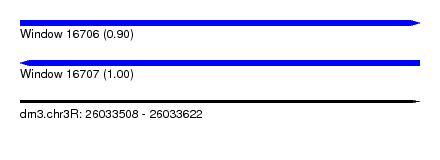
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,033,508 – 26,033,622 |
| Length | 114 |
| Max. P | 0.997501 |

| Location | 26,033,508 – 26,033,622 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 129 |
| Reading direction | forward |
| Mean pairwise identity | 72.74 |
| Shannon entropy | 0.54501 |
| G+C content | 0.43860 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -17.23 |
| Energy contribution | -16.15 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26033508 114 + 27905053 CGGCUCAACAUUGUGGAGUGAUAAGCUGCC----AUUAA-UGUACGUUCUGUUUGACAAUCACUGGCUUUGGA-AGGACAGCCAGCGAAAAACACAAGAACGAUGUAUCAAAAGACCAAC--------- .((.((.((((((((((((.....))).))----).)))-))).((((((...((....((.((((((..(..-....))))))).))....))..))))))...........))))...--------- ( -30.20, z-score = -1.67, R) >droSim1.chr3R 25701459 111 + 27517382 CGGCUCAACAUUGUGGAGUGAUAAGCUGCC----AUUAA-UGUACGUUCUGUUUGACAAUCACUGGCUUUGAA-AGGACAGCCAGCGAAAAACACAAGAACGAUGUAUC---AGACCAAC--------- .((.((.((((((((((((.....))).))----).)))-))).((((((...((....((.((((((((...-.))..)))))).))....))..)))))).......---.))))...--------- ( -30.40, z-score = -1.87, R) >droSec1.super_4 4885462 111 + 6179234 CGGCACAACAUUGUGGAGUGAUAAGCUGCC----AUUAA-UGUACGUUCUGUUUGACAAUCACUGGCUUUGAA-AGGACAGCCAGCGAAAAACACAAGAACGAUGUAUC---AGACCAAC--------- .(..(((((((((((((((.....))).))----).)))-))).((((((...((....((.((((((((...-.))..)))))).))....))..)))))).)))..)---........--------- ( -29.90, z-score = -1.84, R) >droYak2.chr3R 26966779 111 + 28832112 GGGCUCAACAUUGUGGAGUGAAAAGCUGCC----AUUAA-UUUGCGUUCUGUUUGACAAUCCCUGGUUUUGGU-AGGACAACCAGCGAAAAACACAAGAACGGUGUAUC---AGACCAAG--------- ((((.(((....(((((((.....))).))----))...-.))).)))).((((((...((.(((((..((..-....))))))).))...((((.......)))).))---))))....--------- ( -26.00, z-score = 0.40, R) >droEre2.scaffold_4820 8505681 110 - 10470090 AGGCUCAACAUUGUGGAGUGAUA-GCUGCC----AUUAA-UUUACGUUCUGUUUGACAAUCACUGGUUUUGGU-GAGACAACCAGCGAAAAACACAAGAACGGUGUAUC---AGAUCAAC--------- .......((((.(((((((....-))).))----))...-....((((((...((....((.(((((..((..-....))))))).))....))..))))))))))...---........--------- ( -22.70, z-score = 0.75, R) >droAna3.scaffold_13340 21068797 113 + 23697760 CGGCUCAACAUUAUGGAGUGAUUAGCUGCCU----UUAG-UCUUCGUUCUUUUUGACAAUCACUGGUCUUGAGGAGAUUAUCCAGCAAAUAACACAAGAACGGACGGAU---AGACCAGAA-------- (((((...((((....))))...)))))..(----((.(-(((((((((((((((....(((.......)))(((.....)))...........)))))).))))))..---)))).))).-------- ( -22.40, z-score = 0.76, R) >dp4.chr2 29319804 116 - 30794189 CGGCUAAUCAUUGUGGAGUGAGUAGUUGCCUGGGAUUAG-UGUACGUUCUGUUUGACAAUCGCUGGACUAGAA-GGUAAGUCUAGCGAAUAACACAAGAACGGUCAAGG---GUACCAGAC-------- ((((((.(((((....))))).)))))).((((.(((..-((..((((((...((....((((((((((....-....))))))))))....))..))))))..))..)---)).))))..-------- ( -40.60, z-score = -3.64, R) >droPer1.super_7 168605 116 - 4445127 CGGCUAAUCAUUGUGGAGUGAGUAGUUGCCUGGGAUUAG-UGUACGUUCUGUUUGACAAUCGCUGGACUAGAA-GGUAAGUCUAGCGAAUAACACAAGAACGGUCAAGG---GUACCAGAC-------- ((((((.(((((....))))).)))))).((((.(((..-((..((((((...((....((((((((((....-....))))))))))....))..))))))..))..)---)).))))..-------- ( -40.60, z-score = -3.64, R) >droWil1.scaffold_181130 9500521 116 + 16660200 GAGCUUAACAUCGUGGAGUAA-----UGCU-----UUAU-UCUGCGUUCUGUUUGACAAUCGCUGAACUAGGUAAAUGAGUUCAGCGAAUAACACAAGAACGGUCAAUU--GAUAUCAAGGUAAAUGAA ..((((...((((..((((((-----....-----))))-))..)(((((...((....((((((((((.........))))))))))....))..)))))........--)))....))))....... ( -31.00, z-score = -2.20, R) >droVir3.scaffold_12855 3116422 102 + 10161210 AGGCUCUGCAAUGUGGAGUAAACUG--GCCUGGAGUUCGCUUUGCGUUCUGUUUGACAAUCGCUGAACGAGAUUAACAGAUUCAGCGAAUAACACAAGAACGGU------------------------- ............(..(((..((((.--......))))..)))..)(((((...((....(((((((((..........).))))))))....))..)))))...------------------------- ( -26.40, z-score = -0.23, R) >consensus CGGCUCAACAUUGUGGAGUGAUAAGCUGCC____AUUAA_UGUACGUUCUGUUUGACAAUCACUGGACUUGAA_AGGACAUCCAGCGAAAAACACAAGAACGGUGUAUC___AGACCAAC_________ .(((.......................)))..............((((((...((....((.((((((...........)))))).))....))..))))))........................... (-17.23 = -16.15 + -1.08)

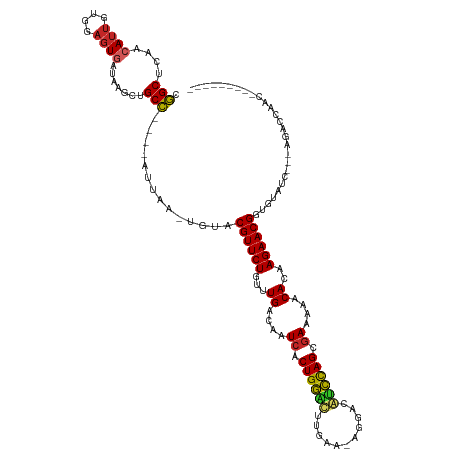
| Location | 26,033,508 – 26,033,622 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 129 |
| Reading direction | reverse |
| Mean pairwise identity | 72.74 |
| Shannon entropy | 0.54501 |
| G+C content | 0.43860 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -23.07 |
| Energy contribution | -21.70 |
| Covariance contribution | -1.37 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.997501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26033508 114 - 27905053 ---------GUUGGUCUUUUGAUACAUCGUUCUUGUGUUUUUCGCUGGCUGUCCU-UCCAAAGCCAGUGAUUGUCAAACAGAACGUACA-UUAAU----GGCAGCUUAUCACUCCACAAUGUUGAGCCG ---------....(((..(((((....((((((..((....(((((((((.....-.....)))))))))....))...))))))...)-)))).----))).(((((.((........)).))))).. ( -31.80, z-score = -2.37, R) >droSim1.chr3R 25701459 111 - 27517382 ---------GUUGGUCU---GAUACAUCGUUCUUGUGUUUUUCGCUGGCUGUCCU-UUCAAAGCCAGUGAUUGUCAAACAGAACGUACA-UUAAU----GGCAGCUUAUCACUCCACAAUGUUGAGCCG ---------...(((..---((((...((((((..((....(((((((((.....-.....)))))))))....))...))))))....-....(----((.((.......)))))...))))..))). ( -30.50, z-score = -1.89, R) >droSec1.super_4 4885462 111 - 6179234 ---------GUUGGUCU---GAUACAUCGUUCUUGUGUUUUUCGCUGGCUGUCCU-UUCAAAGCCAGUGAUUGUCAAACAGAACGUACA-UUAAU----GGCAGCUUAUCACUCCACAAUGUUGUGCCG ---------...(((..---((((...((((((..((....(((((((((.....-.....)))))))))....))...))))))....-....(----((.((.......)))))...))))..))). ( -29.40, z-score = -1.61, R) >droYak2.chr3R 26966779 111 - 28832112 ---------CUUGGUCU---GAUACACCGUUCUUGUGUUUUUCGCUGGUUGUCCU-ACCAAAACCAGGGAUUGUCAAACAGAACGCAAA-UUAAU----GGCAGCUUUUCACUCCACAAUGUUGAGCCC ---------...(((..---((((...((((((..((....((.((((((.....-.....)))))).))....))...))))))....-....(----((.((.......)))))...))))..))). ( -25.00, z-score = -0.04, R) >droEre2.scaffold_4820 8505681 110 + 10470090 ---------GUUGAUCU---GAUACACCGUUCUUGUGUUUUUCGCUGGUUGUCUC-ACCAAAACCAGUGAUUGUCAAACAGAACGUAAA-UUAAU----GGCAGC-UAUCACUCCACAAUGUUGAGCCU ---------((((....---.......((((((..((....(((((((((.....-.....)))))))))....))...))))))....-.....----..))))-.....(((.((...)).)))... ( -24.65, z-score = -0.71, R) >droAna3.scaffold_13340 21068797 113 - 23697760 --------UUCUGGUCU---AUCCGUCCGUUCUUGUGUUAUUUGCUGGAUAAUCUCCUCAAGACCAGUGAUUGUCAAAAAGAACGAAGA-CUAA----AGGCAGCUAAUCACUCCAUAAUGUUGAGCCG --------.....((((---....((((((((((.((.((.(..((((....(((.....)))))))..).)).))..)))))))..))-)...----)))).(((...((........))...))).. ( -23.30, z-score = -0.38, R) >dp4.chr2 29319804 116 + 30794189 --------GUCUGGUAC---CCUUGACCGUUCUUGUGUUAUUCGCUAGACUUACC-UUCUAGUCCAGCGAUUGUCAAACAGAACGUACA-CUAAUCCCAGGCAACUACUCACUCCACAAUGAUUAGCCG --------((((((...---...((..((((((..((.((.(((((.((((....-....)))).))))).)).))...))))))..))-......))))))..(((.(((........))).)))... ( -32.00, z-score = -3.95, R) >droPer1.super_7 168605 116 + 4445127 --------GUCUGGUAC---CCUUGACCGUUCUUGUGUUAUUCGCUAGACUUACC-UUCUAGUCCAGCGAUUGUCAAACAGAACGUACA-CUAAUCCCAGGCAACUACUCACUCCACAAUGAUUAGCCG --------((((((...---...((..((((((..((.((.(((((.((((....-....)))).))))).)).))...))))))..))-......))))))..(((.(((........))).)))... ( -32.00, z-score = -3.95, R) >droWil1.scaffold_181130 9500521 116 - 16660200 UUCAUUUACCUUGAUAUC--AAUUGACCGUUCUUGUGUUAUUCGCUGAACUCAUUUACCUAGUUCAGCGAUUGUCAAACAGAACGCAGA-AUAA-----AGCA-----UUACUCCACGAUGUUAAGCUC .........(((((((((--........(((((..((.((.((((((((((.........)))))))))).)).))...)))))((...-....-----.)).-----.........)))))))))... ( -32.00, z-score = -4.51, R) >droVir3.scaffold_12855 3116422 102 - 10161210 -------------------------ACCGUUCUUGUGUUAUUCGCUGAAUCUGUUAAUCUCGUUCAGCGAUUGUCAAACAGAACGCAAAGCGAACUCCAGGC--CAGUUUACUCCACAUUGCAGAGCCU -------------------------...(((((..((.((.(((((((((..(.....)..))))))))).)).))...)))))((((((..((((......--.))))..)).....))))....... ( -25.40, z-score = -1.53, R) >consensus _________GUUGGUCU___GAUACACCGUUCUUGUGUUAUUCGCUGGAUGUCCU_UUCAAAUCCAGUGAUUGUCAAACAGAACGUACA_UUAAU____GGCAGCUUAUCACUCCACAAUGUUGAGCCG ...........................((((((..((....(((((((((...........)))))))))....))...))))))..............(((.......................))). (-23.07 = -21.70 + -1.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:42 2011