| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,033,256 – 26,033,352 |
| Length | 96 |
| Max. P | 0.966825 |

| Location | 26,033,256 – 26,033,352 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 66.63 |
| Shannon entropy | 0.73184 |
| G+C content | 0.43281 |
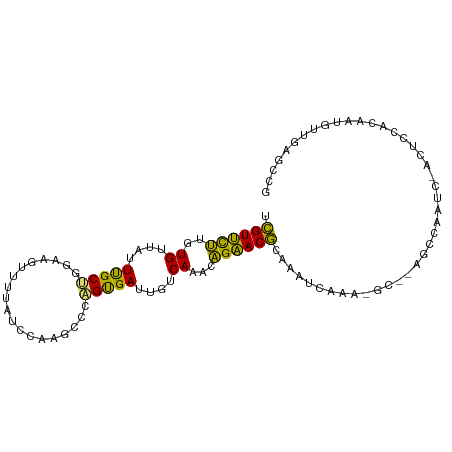
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -10.49 |
| Energy contribution | -9.09 |
| Covariance contribution | -1.39 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
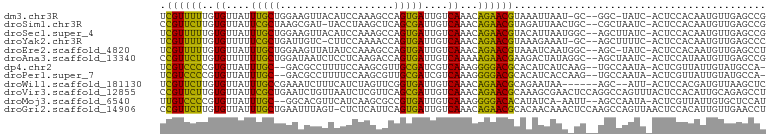
>dm3.chr3R 26033256 96 - 27905053 UCGUUUUUGUGUUAUUUGCUGGAAGUUACAUCCAAAGCCAGUGAUUGUCAAACAGAACGUAAAUUAAU-GC--GGC-UAUC-ACUCCACAAUGUUGAGCCG .((((((..((.((.(..((((...((......))..))))..).)).))...)))))).........-.(--(((-(...-((........))..))))) ( -20.10, z-score = -0.39, R) >droSim1.chr3R 25701119 97 - 27517382 CCGUUCUUGUGUUAUUCGCUAAGCGAU-UACCUAAGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACUGC--CGCUAAUC-ACUCCACAAUGUUGAGCCG .((((((..((.((.(((((.(((...-.......))).))))).)).))...))))))..(((((.....--...)))))-.(((.((...)).)))... ( -22.20, z-score = -1.48, R) >droSec1.super_4 4885223 98 - 6179234 UCGUUUUUGUGUUAUUUGCUGGAAGUUACAUCCAAAGCCAGUGAUUGUCAAACAGAACGUACAUUAAUGGC--AGCUUAUC-ACUCCACAAUGUUGAGCCG .((((((..((.((.(..((((...((......))..))))..).)).))...))))))............--.(((((.(-(........)).))))).. ( -17.10, z-score = 0.89, R) >droYak2.chr3R 26966536 96 - 28832112 UCGUUUUUGUGUUUUUCGCUGAUUGUC-CUUCCAAAACCAGUGAUUGUCAAACAGAACGUAAAGAAAU-GC--AGCUUUUC-ACUCCACAAUGUUGAGCCC .((((((..((....((((((.(((..-....)))...))))))....))...)))))).........-..--.((((..(-(........))..)))).. ( -15.80, z-score = 0.24, R) >droEre2.scaffold_4820 8505418 97 + 10470090 UCGUUUUUGUGUUAUUUGCUGGAAGUUAUAUCCAAAGCCAGUGAUUGUCAAACAGAACGUAAAUCAAUGGC--AGC-UAUC-ACUCCACAAUGUUGAGCCU (((...(((((......((((((.......)))...((((.(((((.((.....)).....))))).))))--)))-....-....)))))...))).... ( -20.04, z-score = -0.41, R) >droAna3.scaffold_13340 21068577 98 - 23697760 CCGUUCUUGUGUUUUUUGCUGGAUAAUCUCCUCAAGACCAGUGAUUGUCAAAAAGAACGAAGACUAUAGGC--AGCUAAUC-ACUCCAUAAUGUUGAGCCG .(((((((.((....(..((((....(((.....)))))))..)....))..))))))).........(((--(((.....-..........)))..))). ( -19.46, z-score = -0.31, R) >dp4.chr2 29319962 95 + 30794189 UCGUCCCCGUGUUAUUUGC--GACGCCUUUUCCAAGCGUUGCGAUCGUCAAAGGGGACGCACAUCAUCAAG--UGCCAAUA-ACUCGUUAUUGUAUGCCA- ..((((((.((....((((--(((((.........)))))))))....))..))))))((((........)--)))(((((-(....)))))).......- ( -33.70, z-score = -4.00, R) >droPer1.super_7 168763 95 + 4445127 UCGUCCCCGUGUUAUUUGC--GACGCCUUUUCCAAGCGUUGCGAUCGUCAAAGGGGACGCACAUCACCAAG--UGCCAAUA-ACUCGUUAUUGUAUGCCA- ..((((((.((....((((--(((((.........)))))))))....))..))))))((((........)--)))(((((-(....)))))).......- ( -33.70, z-score = -3.91, R) >droWil1.scaffold_181130 9500252 92 - 16660200 UCGUUCUUGUGUUAUUUGCCGAAAUCUUUCAUCUAGUUCGGUGAUUGUCAAACAGAACGCAGAAUAA------AGC--AUU-ACUCCACGAUGUUAAGCUC .((((((..((.((.(..(((((..((.......)))))))..).)).))...))))))........------(((--(((-.......))))))...... ( -17.80, z-score = -0.46, R) >droVir3.scaffold_12855 3116422 101 - 10161210 CCGUUCUUGUGUUAUUCGCUGAAUCUGUUAAUCUCGUUCAGCGAUUGUCAAACAGAACGCAAAGCGAACUCCAGGCCAGUUUACUCCACAUUGCAGAGCCU ..(((((..((.((.(((((((((..(.....)..))))))))).)).))...)))))((((((..((((.......))))..)).....))))....... ( -25.40, z-score = -1.56, R) >droMoj3.scaffold_6540 24062899 95 + 34148556 UUGUCCCCGUGUUAUUUGC--GGCACGUUCAUCAAGCGCCGUGAUUGUCAAAGGGGACACAUAUCA-AAUU--AGCCAAUA-ACUCGUUAUUGUGCUCCAU .(((((((.((.((.(..(--(((.(.........).))))..).)).))..))))))).......-....--((((((((-(....)))))).))).... ( -31.30, z-score = -3.60, R) >droGri2.scaffold_14906 5812972 100 - 14172833 CCGUUCUUGUGUUAUUUGCUGAAUUUAGU-CUCUCAUUCAGUGAUUGUCAAACAGAACGCACAACAAACUCCAAGCCAGUUAACUCCACAUUGUUGAACCU .((((((..((.((.(..((((((.....-.....))))))..).)).))...))))))..((((((......((........)).....))))))..... ( -17.90, z-score = -0.67, R) >consensus UCGUUCUUGUGUUAUUUGCUGGAAGUUUUAUCCAAGCCCAGUGAUUGUCAAACAGAACGCAAAUCAAA_GC__AGCCAAUC_ACUCCACAAUGUUGAGCCG .((((((..((....(((((...................)))))....))...)))))).......................................... (-10.49 = -9.09 + -1.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:41 2011