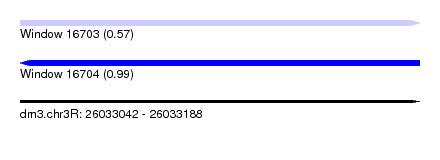
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,033,042 – 26,033,188 |
| Length | 146 |
| Max. P | 0.994564 |

| Location | 26,033,042 – 26,033,188 |
|---|---|
| Length | 146 |
| Sequences | 12 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 67.52 |
| Shannon entropy | 0.66932 |
| G+C content | 0.42057 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -14.76 |
| Energy contribution | -13.97 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
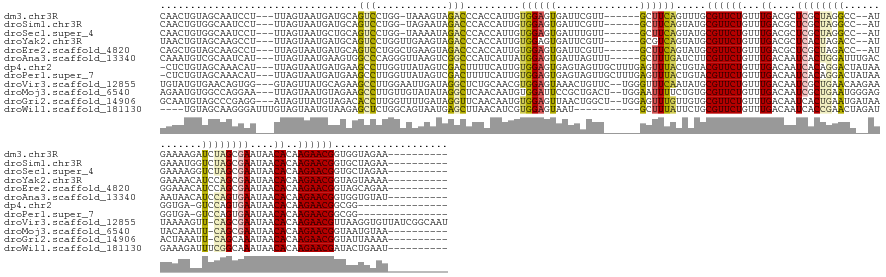
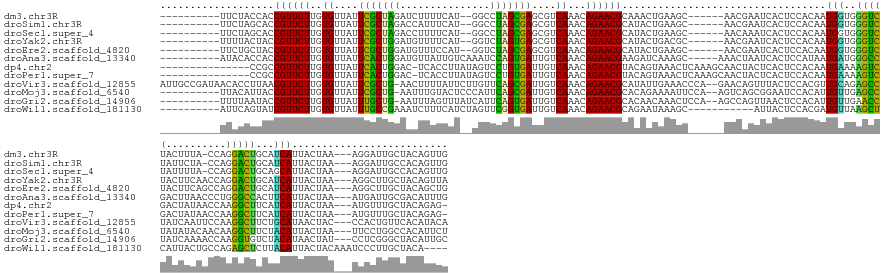
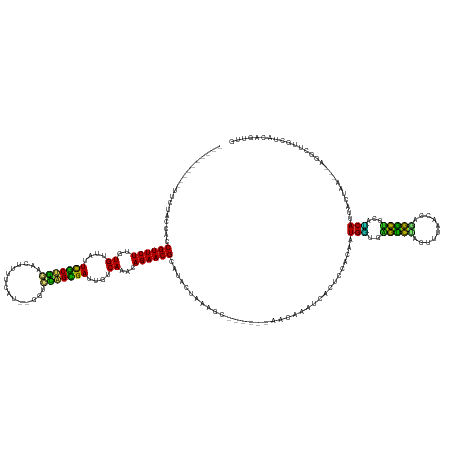
>dm3.chr3R 26033042 146 + 27905053 CAACUGUAGCAAUCCU---UUAGUAAUGAUGCAGUCCUGG-UAAAGUAGACCCACCAUUGUGGAGUGAUUCGUU------GCUUCAGUUUGCGUUCUGUUUGACGCUCGCUAGGCC--AUGAAAAGAUCUAGCGAAUAACACAAGAACGGUGGUAGAA---------- ..((..(.((((..((---..((((((((..((.((((((-(...........))))....))).))..)))))------)))..)).))))(((((...((....(((((((..(--.......)..)))))))....))..))))).)..))....---------- ( -40.40, z-score = -0.96, R) >droSim1.chr3R 25701245 146 + 27517382 CAACUGUGGCAAUCCU---UUAGUAAUGAUGCAGUCCUGG-UAGAAUAGACCCACCAUUGUGGAGUGAUUCGUU------GCUUCAGUAUGCGUUCUGUUUGACGCUCGCUAGGCC--AUGAAAUGGUCUAGCGAAUAACACAAGAACGGUGCUAGAA---------- ....(.(((((...((---..((((((((..((.((((((-(.(.......).))))....))).))..)))))------)))..))....((((((...((....((((((((((--((...))))))))))))....))..)))))).))))).).---------- ( -48.80, z-score = -2.65, R) >droSec1.super_4 4885009 146 + 6179234 CAACUGUGGCAAUCCU---UUAGUAAUGCUGCAGUCCUGG-UAAAAUAGACCCACCAUUGUGGAGUGAUUUGUU------GCUUCAGUAUGCGUUCUGUUUGACGCUCGCUAGGCC--AUGAAAAGGUCUAGCGAAUAACACAAGAACGGUGCUAGAA---------- ..((((..(((.....---.......)))..)))).((((-((............(((..(((((..(....).------.)))))..)))((((((...((....((((((((((--.......))))))))))....))..)))))).))))))..---------- ( -48.00, z-score = -2.55, R) >droYak2.chr3R 26966321 147 + 28832112 UAACUGUAGCAAGCCU---UUAGUAAUGAUGCAGUCCUGGUUGAAGUAGACCCACCAUUGUGGAGUGAUUCGUU------GCGUCAGUAUGCGUUCUGUUUGACGCUCACUAGACC--AUGAAAACAUCCAGCGAAUAACACAAGAACGGUAGUAAAA---------- ..(((((.....((..---(((....))).))..((((((.((......((((((....)))).))..(((((.------(.((((((..(((((......)))))..))).))))--)))))..)).)))).))...........))))).......---------- ( -34.30, z-score = 0.18, R) >droEre2.scaffold_4820 8505196 147 - 10470090 CAGCUGUAGCAAGCCU---UUAGUAAUGAUGCAGUCCUGGCUGAAGUAGACCCACCAUUGUGGAGUGAUUCGUU------GCUUCAGUAUGCGUUCUGUUUGACGCUCGCUAGACC--AUGGAAACAUCCAGCGAAUAACACAAGAACGGUAGCAGAA---------- ..((((..(((.((..---..((((((((..((((....))))......((((((....)))).))...)))))------)))...)).)))(((((...((....(((((.((..--.((....)))).)))))....))..)))))..))))....---------- ( -41.60, z-score = -0.51, R) >droAna3.scaffold_13340 21068334 150 + 23697760 CAAAUGUCGCAAUCAU---UUAGUAAUGAAGUGGCCCAGGGUUAAGUCGGCCCAUCAUUAUGGAGUGAUUAGUUU-----GCUUUGAUCUUCGUUCUGUUUGACAAUCACUGGAUUUGACAAUAACAUCCAGUGAAUAACACAAGAACGGUGGUGUAU---------- ...((..(((((((((---((.(((((((.(.((((............))))).))))))).)))))))).....-----...........((((((...((....(((((((((.(.......).)))))))))....))..)))))))))..))..---------- ( -44.90, z-score = -2.48, R) >dp4.chr2 29319579 148 - 30794189 -CUCUGUAGCAAACAU---UUAGUAAUGAUGAAGCCUUGGUUAUAGUCGACUUUUCAUUGUGGAGUGAGUAGUUGCUUUGAGUUUACUGUACGUUCUGUUUGACAAUCACAGGACUAUAAGGUGA-GUCCAGUGAAUAACACAAGAACGGCGG--------------- -(((...(((((..((---(((.(.(..((((((..((((......))))..))))))..)..).)))))..)))))..)))....((((.((((((...((....((((.(((((........)-)))).))))....))..))))))))))--------------- ( -41.20, z-score = -1.32, R) >droPer1.super_7 168380 148 - 4445127 -CUCUGUAGCAAACAU---UUAGUAAUGAUGAAGCCUUGGUUAUAGUCGACUUUUCAUUGUGGAGUGAGUAGUUGCUUUGAGUUUACUGUACGUUCUGUUUGACAAUCACAGGACUAUAAGGUGA-GUCCAGUGAAUAACACAAGAACGGCGG--------------- -(((...(((((..((---(((.(.(..((((((..((((......))))..))))))..)..).)))))..)))))..)))....((((.((((((...((....((((.(((((........)-)))).))))....))..))))))))))--------------- ( -41.20, z-score = -1.32, R) >droVir3.scaffold_12855 3116184 162 + 10161210 UGUAUGUGAACAGUGG---GUAGUUAUGCAGAAGCCUUGGAAUUGAUAGGCUCUGCAACGUGGAGUAAACUGUUC--UGGGUUUCAAUAUGCGUUCUGUUUGACAAUCGCUGAACAAGAAUAAAAGUU-CAGCGAAUAACACAAGAACGUUAAGGUGUUAUCGGCAAU ....((((((((((..---.((.((.((((((.((((..........)))))))))).....)).)).)))))))--..(((..((....(((((((...((....(((((((((..........)))-))))))....))..))))))).....))..))).))).. ( -50.40, z-score = -3.11, R) >droMoj3.scaffold_6540 24062692 152 - 34148556 AGAAUGUGGCCAGGAA---UUAGUAAUGUAGAAGCCUUGUUGUAUAUAGGCUCAACAAUGUGGAUUCCGCUGACU--UGGAAUUUUCUGUGCGUUCUGUUUGACAAUCGCUGAAUGGGAGUACAAAUU-CAGCGAAUAACACAAGAACGGUAAUGUAA---------- ((((.((..(((((..---.......(((.((.((((..........)))))).)))..((((...))))...))--))).)).)))).((((((((((((.....(((((((((..(....)..)))-)))))).........)))))).)))))).---------- ( -40.84, z-score = -1.34, R) >droGri2.scaffold_14906 5812528 152 + 14172833 GCAAUGUAGCCCGAGG---AUAGUUAUGUAGACACCUUGGUUUUGAUAGGUUCAACAAUGUGGAGUUAACUGGCU--UGGAGUUUGUUGUGCGUUCUGUUUGACAAUCACUGAAUGAUAAACUAAAUU-CAGCAAAUAACACAAGAACGGUAUUAAAA---------- ((((((.((((((((.---.((((((......(((.(((...((((.....))))))).)))....)))))).))--))).))))))))).((((((...((.......((((((..........)))-))).......))..)))))).........---------- ( -33.54, z-score = 0.17, R) >droWil1.scaffold_181130 9499940 143 + 16660200 ----UGUAGCAAGGGAUUUGUAGUAAUGUAAGAGCUCUGGCAGUAAUGAGCUUAACAUCGUGGAGUAAU-----------GCUUUAUUCUGCGUUCUGUUUGACAAUCACCGAACUAGAUGAAAGAUUUCGGCAAAUAACACAAGAACGAUACUGAAU---------- ----........(..((((((...(((.(..((((((..........))))))..(((((..((((((.-----------...))))))..).....(((((........)))))..))))..).)))...))))))..)..................---------- ( -30.20, z-score = 0.38, R) >consensus CAACUGUAGCAAGCCU___UUAGUAAUGAUGAAGCCCUGGUUAAAAUAGACCCACCAUUGUGGAGUGAUUCGUU______GCUUCAGUAUGCGUUCUGUUUGACAAUCACUGGACC__AUGAAAAGAUCCAGCGAAUAACACAAGAACGGUAGUAGAA__________ .................................(((............))).........((((((..............)))))).....((((((...((....((((((((.............))))))))....))..))))))................... (-14.76 = -13.97 + -0.79)
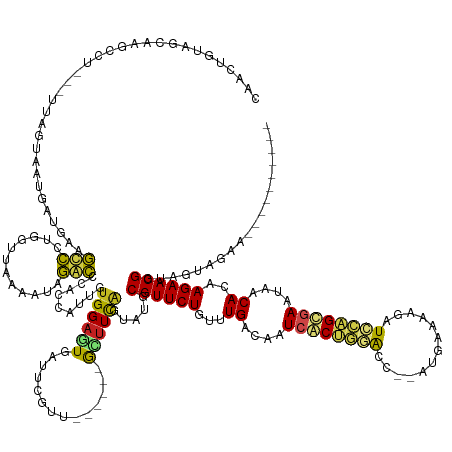
| Location | 26,033,042 – 26,033,188 |
|---|---|
| Length | 146 |
| Sequences | 12 |
| Columns | 168 |
| Reading direction | reverse |
| Mean pairwise identity | 67.52 |
| Shannon entropy | 0.66932 |
| G+C content | 0.42057 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -20.81 |
| Energy contribution | -18.64 |
| Covariance contribution | -2.17 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26033042 146 - 27905053 ----------UUCUACCACCGUUCUUGUGUUAUUCGCUAGAUCUUUUCAU--GGCCUAGCGAGCGUCAAACAGAACGCAAACUGAAGC------AACGAAUCACUCCACAAUGGUGGGUCUACUUUA-CCAGGACUGCAUCAUUACUAA---AGGAUUGCUACAGUUG ----------.........((((((..((...((((((((..(.......--)..))))))))...))...))))))..(((((.(((------(....(((.......((((((((((((......-...))))).))))))).....---..))))))).))))). ( -40.64, z-score = -2.45, R) >droSim1.chr3R 25701245 146 - 27517382 ----------UUCUAGCACCGUUCUUGUGUUAUUCGCUAGACCAUUUCAU--GGCCUAGCGAGCGUCAAACAGAACGCAUACUGAAGC------AACGAAUCACUCCACAAUGGUGGGUCUAUUCUA-CCAGGACUGCAUCAUUACUAA---AGGAUUGCCACAGUUG ----------.........((((((..((...((((((((.((((...))--)).))))))))...))...))))))...((((..((------(....(((.......((((((((((((......-...))))).))))))).....---..))))))..)))).. ( -38.94, z-score = -1.48, R) >droSec1.super_4 4885009 146 - 6179234 ----------UUCUAGCACCGUUCUUGUGUUAUUCGCUAGACCUUUUCAU--GGCCUAGCGAGCGUCAAACAGAACGCAUACUGAAGC------AACAAAUCACUCCACAAUGGUGGGUCUAUUUUA-CCAGGACUGCAGCAUUACUAA---AGGAUUGCCACAGUUG ----------......((((((...((((....(((((((.((.......--)).)))))))((((........))))..........------............))))))))))(((.......)-))..(((((..(((...(...---.)...)))..))))). ( -37.40, z-score = -0.96, R) >droYak2.chr3R 26966321 147 - 28832112 ----------UUUUACUACCGUUCUUGUGUUAUUCGCUGGAUGUUUUCAU--GGUCUAGUGAGCGUCAAACAGAACGCAUACUGACGC------AACGAAUCACUCCACAAUGGUGGGUCUACUUCAACCAGGACUGCAUCAUUACUAA---AGGCUUGCUACAGUUA ----------........((((((((((((((.(((((((((........--.)))))))))((((........))))....))))))------)).))))........((((((((((((..........))))).))))))).....---.))............. ( -35.70, z-score = -0.33, R) >droEre2.scaffold_4820 8505196 147 + 10470090 ----------UUCUGCUACCGUUCUUGUGUUAUUCGCUGGAUGUUUCCAU--GGUCUAGCGAGCGUCAAACAGAACGCAUACUGAAGC------AACGAAUCACUCCACAAUGGUGGGUCUACUUCAGCCAGGACUGCAUCAUUACUAA---AGGCUUGCUACAGCUG ----------...(((..(((((((..((...((((((((((........--.))))))))))...))...))))).....((((((.------...((..((((.......))))..))..))))))...))...)))..........---.((((......)))). ( -36.80, z-score = 0.42, R) >droAna3.scaffold_13340 21068334 150 - 23697760 ----------AUACACCACCGUUCUUGUGUUAUUCACUGGAUGUUAUUGUCAAAUCCAGUGAUUGUCAAACAGAACGAAGAUCAAAGC-----AAACUAAUCACUCCAUAAUGAUGGGCCGACUUAACCCUGGGCCACUUCAUUACUAA---AUGAUUGCGACAUUUG ----------.........((((((..((.((.(((((((((...........))))))))).)).))...)))))).....((((..-----....((((((.....((((((..((((.(........).))))...))))))....---.)))))).....)))) ( -38.50, z-score = -2.59, R) >dp4.chr2 29319579 148 + 30794189 ---------------CCGCCGUUCUUGUGUUAUUCACUGGAC-UCACCUUAUAGUCCUGUGAUUGUCAAACAGAACGUACAGUAAACUCAAAGCAACUACUCACUCCACAAUGAAAAGUCGACUAUAACCAAGGCUUCAUCAUUACUAA---AUGUUUGCUACAGAG- ---------------....((((((..((.((.((((.((((-(........))))).)))).)).))...)))))).........(((..(((((.............(((((.(((((............)))))..)))))((...---..)))))))...)))- ( -30.20, z-score = -1.19, R) >droPer1.super_7 168380 148 + 4445127 ---------------CCGCCGUUCUUGUGUUAUUCACUGGAC-UCACCUUAUAGUCCUGUGAUUGUCAAACAGAACGUACAGUAAACUCAAAGCAACUACUCACUCCACAAUGAAAAGUCGACUAUAACCAAGGCUUCAUCAUUACUAA---AUGUUUGCUACAGAG- ---------------....((((((..((.((.((((.((((-(........))))).)))).)).))...)))))).........(((..(((((.............(((((.(((((............)))))..)))))((...---..)))))))...)))- ( -30.20, z-score = -1.19, R) >droVir3.scaffold_12855 3116184 162 - 10161210 AUUGCCGAUAACACCUUAACGUUCUUGUGUUAUUCGCUG-AACUUUUAUUCUUGUUCAGCGAUUGUCAAACAGAACGCAUAUUGAAACCCA--GAACAGUUUACUCCACGUUGCAGAGCCUAUCAAUUCCAAGGCUUCUGCAUAACUAC---CCACUGUUCACAUACA ...................((((((..((.((.((((((-(((..........))))))))).)).))...))))))..............--(((((((...........((((((((((..........)))).)))))).......---..)))))))....... ( -44.55, z-score = -5.24, R) >droMoj3.scaffold_6540 24062692 152 + 34148556 ----------UUACAUUACCGUUCUUGUGUUAUUCGCUG-AAUUUGUACUCCCAUUCAGCGAUUGUCAAACAGAACGCACAGAAAAUUCCA--AGUCAGCGGAAUCCACAUUGUUGAGCCUAUAUACAACAAGGCUUCUACAUUACUAA---UUCCUGGCCACAUUCU ----------.........((((((..((.((.((((((-(((..(....)..))))))))).)).))...))))))..............--.((((..(((((......(((.((((((..........))))))..)))......)---))))))))........ ( -35.30, z-score = -2.85, R) >droGri2.scaffold_14906 5812528 152 - 14172833 ----------UUUUAAUACCGUUCUUGUGUUAUUUGCUG-AAUUUAGUUUAUCAUUCAGUGAUUGUCAAACAGAACGCACAACAAACUCCA--AGCCAGUUAACUCCACAUUGUUGAACCUAUCAAAACCAAGGUGUCUACAUAACUAU---CCUCGGGCUACAUUGC ----------.........((((((..((.((.(..(((-(((..........))))))..).)).))...))))))..............--((((..(((((........)))))((((..........))))..............---.....))))....... ( -25.80, z-score = 0.05, R) >droWil1.scaffold_181130 9499940 143 - 16660200 ----------AUUCAGUAUCGUUCUUGUGUUAUUUGCCGAAAUCUUUCAUCUAGUUCGGUGAUUGUCAAACAGAACGCAGAAUAAAGC-----------AUUACUCCACGAUGUUAAGCUCAUUACUGCCAGAGCUCUUACAUUACUACAAAUCCCUUGCUACA---- ----------((((.....((((((..((.((.(..(((((..((.......)))))))..).)).))...))))))..))))..(((-----------(.........(((((..(((((..........)))))...))))).............))))...---- ( -26.45, z-score = -0.56, R) >consensus __________UUCUACCACCGUUCUUGUGUUAUUCGCUGGAACUUUUCAU__GGUCCAGCGAUUGUCAAACAGAACGCAUACUAAAGC______AACAAAUCACUCCACAAUGGUGAGUCUACUUUAACCAGGGCUGCAUCAUUACUAA___AGGCUUGCUACAGUUG ...................((((((..((....(((((((...............)))))))....))...))))))..................................(((..(((((..........)))))...))).......................... (-20.81 = -18.64 + -2.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:40 2011