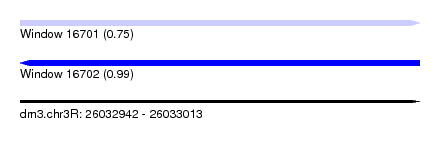
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,032,942 – 26,033,013 |
| Length | 71 |
| Max. P | 0.994730 |

| Location | 26,032,942 – 26,033,013 |
|---|---|
| Length | 71 |
| Sequences | 12 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.56196 |
| G+C content | 0.42217 |
| Mean single sequence MFE | -15.68 |
| Consensus MFE | -8.14 |
| Energy contribution | -7.69 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.751929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
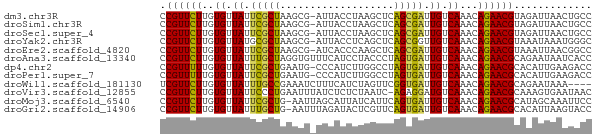
>dm3.chr3R 26032942 71 + 27905053 GGCAGUUAAUCUACGUUCUGUUUGACAAUCGCUGAGCUUAGGUAAU-CGCUUAGCGAAUAACACAAGAACGG .............((((((...((....(((((((((.........-.)))))))))....))..)))))). ( -21.30, z-score = -2.89, R) >droSim1.chr3R 25701145 71 + 27517382 GGCAGUUAAUCUACGUUCUGUUUGACAAUCGCUGAGCUUAGGUAAU-CGCUUAGCGAAUAACACAAGAACGG .............((((((...((....(((((((((.........-.)))))))))....))..)))))). ( -21.30, z-score = -2.89, R) >droSec1.super_4 4884909 71 + 6179234 GGCAGUUAAUCUACGUUCUGUUUGACAAUCGCUGAGCUUAGGUAAU-CGCUUAGCGAAUAACACAAGAACGG .............((((((...((....(((((((((.........-.)))))))))....))..)))))). ( -21.30, z-score = -2.89, R) >droYak2.chr3R 26966220 71 + 28832112 GCCCAUUUAUUUACGUUCUGUUUGACAACCGCUGAGCUGAGGUAAU-CGCUUAGCGCAUAACACAAGAACGG .............((((((...((.....((((((((.((.....)-))))))))).....))..)))))). ( -21.00, z-score = -2.90, R) >droEre2.scaffold_4820 8505095 71 - 10470090 GGCCGUUAAUUUACGUUCUGUUUGACAAUCGCUGAGCUUGGGUGAU-CGCUUAGCGAAUAACACAAGAACGG .............((((((...((....(((((((((.........-.)))))))))....))..)))))). ( -21.30, z-score = -2.25, R) >droAna3.scaffold_13340 21068243 72 + 23697760 GGUGAUUAUUCUGCGUUCUGUUUGACAAUCACUAGGGUAGGAUGAAACACCUAGCAAAUAACACAAGAACGG ((((.(((((((((..((((..((.....)).)))))))))))))..))))..................... ( -13.60, z-score = -0.52, R) >dp4.chr2 29319488 71 - 30794189 GGUCUUCAAUGUGCGUUCUGUUUGACAAUCACUAGGCCAAGAUGGG-CAUUCAGCGAAUAACACAAAAACGG .........((((.(((......)))..((.((..(((......))-)....)).))....))))....... ( -9.00, z-score = 1.67, R) >droPer1.super_7 168289 71 - 4445127 GGUCUUCAAUGUGCGUUCUGUUUGACAAUCACUAGGCCAAGAUGGG-CAUUCAGCGAAUAACACAAAAACGG .........((((.(((......)))..((.((..(((......))-)....)).))....))))....... ( -9.00, z-score = 1.67, R) >droWil1.scaffold_181130 9499202 68 + 16660200 ----UUUAUUCUGCGUUCUGUUUGACAAUCACCGAACUAGAUGAAAGAUUUCGGCAAAUAACACAAGAACGA ----.........((((((...((.......(((((.............))))).......))..)))))). ( -10.36, z-score = -0.30, R) >droVir3.scaffold_12855 3116099 71 + 10161210 GUUAUUCACUUUGCGUUCUGUUUGACAUCCUCU-GAUUAGAGAGAUAAAUUCAGGGAAUAACACAAGAACGG .............((((((........(((.((-((..............)))))))........)))))). ( -14.23, z-score = -0.87, R) >droMoj3.scaffold_6540 24062566 71 - 34148556 GGAAUUUGCUAUGCGUUCUGUUUGACAAUCACUGAAUGAUAAUGCUAAUU-CAGCGAAUAACACAAGAACGG .............((((((...((....((.((((((..........)))-))).))....))..)))))). ( -14.30, z-score = -1.42, R) >droGri2.scaffold_14906 5811812 71 + 14172833 GGUACUUAAUGUGCGUUCUGUUUGACAAUCACUGAACGAGUAUCUAAAUU-CAGCAAAUAACACAAGAACGG .((((.....))))((((((((((........)))))((((......)))-).............))))).. ( -11.50, z-score = -0.26, R) >consensus GGCAUUUAAUCUGCGUUCUGUUUGACAAUCACUGAGCUAAGGUGAU_CGCUCAGCGAAUAACACAAGAACGG .............((((((...((.......(((((.............))))).......))..)))))). ( -8.14 = -7.69 + -0.45)

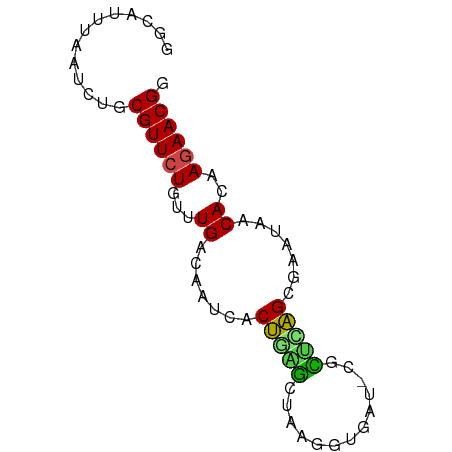
| Location | 26,032,942 – 26,033,013 |
|---|---|
| Length | 71 |
| Sequences | 12 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.56196 |
| G+C content | 0.42217 |
| Mean single sequence MFE | -16.98 |
| Consensus MFE | -10.41 |
| Energy contribution | -9.82 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26032942 71 - 27905053 CCGUUCUUGUGUUAUUCGCUAAGCG-AUUACCUAAGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACUGCC ..(((((..((.((.(((((.(((.-.........))).))))).)).))...)))))((((.....)))). ( -19.80, z-score = -2.95, R) >droSim1.chr3R 25701145 71 - 27517382 CCGUUCUUGUGUUAUUCGCUAAGCG-AUUACCUAAGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACUGCC ..(((((..((.((.(((((.(((.-.........))).))))).)).))...)))))((((.....)))). ( -19.80, z-score = -2.95, R) >droSec1.super_4 4884909 71 - 6179234 CCGUUCUUGUGUUAUUCGCUAAGCG-AUUACCUAAGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACUGCC ..(((((..((.((.(((((.(((.-.........))).))))).)).))...)))))((((.....)))). ( -19.80, z-score = -2.95, R) >droYak2.chr3R 26966220 71 - 28832112 CCGUUCUUGUGUUAUGCGCUAAGCG-AUUACCUCAGCUCAGCGGUUGUCAAACAGAACGUAAAUAAAUGGGC .((((((..((.((..((((.(((.-.........))).))))..)).))...))))))............. ( -16.00, z-score = -0.24, R) >droEre2.scaffold_4820 8505095 71 + 10470090 CCGUUCUUGUGUUAUUCGCUAAGCG-AUCACCCAAGCUCAGCGAUUGUCAAACAGAACGUAAAUUAACGGCC .((((((..((.((.(((((.(((.-.........))).))))).)).))...))))))............. ( -18.90, z-score = -2.68, R) >droAna3.scaffold_13340 21068243 72 - 23697760 CCGUUCUUGUGUUAUUUGCUAGGUGUUUCAUCCUACCCUAGUGAUUGUCAAACAGAACGCAGAAUAAUCACC .((((((..((.((.(..(((((.((........)))))))..).)).))...))))))............. ( -18.50, z-score = -3.04, R) >dp4.chr2 29319488 71 + 30794189 CCGUUUUUGUGUUAUUCGCUGAAUG-CCCAUCUUGGCCUAGUGAUUGUCAAACAGAACGCACAUUGAAGACC .((((((..((.((.((((((...(-((......))).)))))).)).))...))))))............. ( -14.40, z-score = -0.66, R) >droPer1.super_7 168289 71 + 4445127 CCGUUUUUGUGUUAUUCGCUGAAUG-CCCAUCUUGGCCUAGUGAUUGUCAAACAGAACGCACAUUGAAGACC .((((((..((.((.((((((...(-((......))).)))))).)).))...))))))............. ( -14.40, z-score = -0.66, R) >droWil1.scaffold_181130 9499202 68 - 16660200 UCGUUCUUGUGUUAUUUGCCGAAAUCUUUCAUCUAGUUCGGUGAUUGUCAAACAGAACGCAGAAUAAA---- .((((((..((.((.(..(((((..((.......)))))))..).)).))...)))))).........---- ( -15.30, z-score = -1.76, R) >droVir3.scaffold_12855 3116099 71 - 10161210 CCGUUCUUGUGUUAUUCCCUGAAUUUAUCUCUCUAAUC-AGAGGAUGUCAAACAGAACGCAAAGUGAAUAAC .((((((.((.....(((((((..............))-)).)))......))))))))............. ( -13.74, z-score = -1.44, R) >droMoj3.scaffold_6540 24062566 71 + 34148556 CCGUUCUUGUGUUAUUCGCUG-AAUUAGCAUUAUCAUUCAGUGAUUGUCAAACAGAACGCAUAGCAAAUUCC .((((((..((.((.((((((-(((.(......).))))))))).)).))...))))))............. ( -17.80, z-score = -3.08, R) >droGri2.scaffold_14906 5811812 71 - 14172833 CCGUUCUUGUGUUAUUUGCUG-AAUUUAGAUACUCGUUCAGUGAUUGUCAAACAGAACGCACAUUAAGUACC .((((((..((.((.(..(((-(((..........))))))..).)).))...))))))............. ( -15.30, z-score = -1.52, R) >consensus CCGUUCUUGUGUUAUUCGCUAAACG_AUCACCUUAGCUCAGUGAUUGUCAAACAGAACGCAGAUUAAAUACC .((((((..((.((.(((((...................))))).)).))...))))))............. (-10.41 = -9.82 + -0.59)
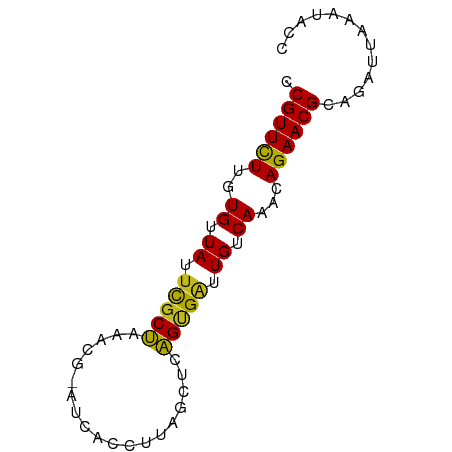
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:38 2011